ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Kiran Kishore V1, Saddam Shaik2, Abdul Mutallib Mulla3, Siva Prasada Reddy Pocha4
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
ABSTRACT:In this project we proposed a novel multifunctional platform focusing on the clinical diagnosis of kidneys and their pathology (tumors, stones and cysts), using a ―templates‖-based technique. As a first step, specialist clinicians train the system by accurately annotating the kidneys and their abnormalities creating ―3-D golden standard models.‖ Then, medical technicians experimentally adjust rules and parameters (stored as ―templates‖) for the integrated ―automatic recognition framework‖ to achieve results which are closest to those of the clinicians. These parameters can later be used by nonexperts to achieve increased automation in the identification process. The system‘s functionality was tested on 20 MRI datasets (552 images), while the ―automatic 3-D models‖ created were validated against the ―3-D golden standard models.‖ Results are promising as they yield an average accuracy of 97.2% in successfully identifying kidneys and 96.1% of their abnormalities thus outperforming existing methods both in accuracy and in processing time needed.
Keywords |
| Abnormalities detection, automatic annotate-on kidney pathology, kidney segmentation, region of interest (ROI), stone, tumor. |
INTRODUCTION |
| The rapid evolution of advanced medical image modalities such as the modern MRI scanners and the large amount of data provided have brought about the need for more automatic processes in computeraided diagnosis. Clinicians need to examine large numbers of complex medical images to detect abnormalities a difficult and time consuming task. Hence, there is a need for systems that will automatically detect organs and their possible abnormalities and provide useful metrics. Several algorithms detect kidney abnormalities addressing the challenge of increased difficulty in their delineation due to their intensity variation. Prevostetal.[1] Had automatically localized the kidney with an oval ellipsoid detector, and then applied deformation of this ellipsoid with a model-based approach in the segmentation process. Using the Dice Similarity Coefficient (DSC) asymmetric [2], this system achieved a DSC of 87.5%. Similar to this platform, they calculated the accuracy of automatic segmentation outcome by comparing it with the result of the semiautomatic segmentation method coming from the radiologist‘s work (golden standard). Lin et al.‘s [3] model-based approach for kidney segmentation achieved an average correlation coefficient of 88%,while [4] used Bayesian concepts for a probability map generation to achieve an automatic kidney Parenchyma volume try with a DSC of 90.3%. [5] Used an automated graph-cuts segmentation technique for dynamic contrast-enhanced 3-D MR venography achieving a DSC of 96% for the kidney and 90% for the cortex and the medulla. Their method was very fast (approximately 20 min) compared with the time needed for a manual segmentation of about 2.5 h, In [6], the authors presented a combination of texture feature and a statistical matching of geometrical shapes of kidneys for an automatic segmentation in3-D MRI images with a mean DSC of 90.6%. Concerning abnormality detection, [7] presented a finite-element method based on 3-D Tumour growth prediction forkidney tumours, with an average true positive fraction of 91.4% on all tumours. Tamilsalvia and Thingraj [8] used an improved seeded region growing method and classification of kidney images with stones and focused on the kidney image segmentation and diagnosis for stone detection. In [8] the authors achieved a DSC of 95%.In [9] three neural network algorithms for diagnosis of kidney stones diseases were tested. They claim that the multi layer perception with two hidden layers and the back propagation algorithm produced the best model for the diagnosis of kidney stone disease with an accuracy of 92%. In [10], an automatic segmentation algorithm for segmenting liver and tumour based on threshold and region growing techniques was used. The tumour was segmented with an alternative FCM clustering algorithm and the DSC for liver and tumour was 95.8% and 89.8%, respectively. The methodology of [11] was closer to the one presented here as their automatic segmentation method was based on association rule-mining to enhance the diagnosis and classification of kidney images. They used conditions and criteria to adjust their system, thus achieving results with an average accuracy of 92%. In [14] a survey of systems focusing on identifying pathologies in transplanted kidneys with most of those methods suffering from low accuracy, or the need for extended involvement of clinicians to identify organs and abnormalities was presented. In the field of transplant kidney rejection, [12] and [13] presented a related system based on dynamically enhancing the contrast of 2-D MRI images with achieve organ identification. The firstpart of their method is related to our own system and achieved an average accuracy of 92.31% (although our system focuses on tumour detection as well as generic abnormalities). This research presents the automatic tumour detection (ATD) platform; an innovative system to support a method for increased automation of kidney detection as well as their abnormalities (tumours, stones, and cysts).` |
| This system uses a novel approach to increase both the accuracy and the automation of the process. Initially, a clinician invokes a fast version of the region growing segmentation algorithm and a set of advanced correction tools to semiautomatically detect and annotate theregionsofinterest(ROI),andtoannotattheorgansandtheirpotential abnormalities (tumours, stones and cysts) in a specific MRI dataset. Next, a medical technician defines the rules and parameters for the automatic recognition framework and thus generates 3-D models of the organs and their potential pathology similar to the ones defined by the clinicians. This produces a template that includes all the sensitivity parameters that govern the dataset that come from a specifically calibrated MRI. The final step is for any user to apply this template‘s settings to all other datasets of the same type and automatically identify the organs and their dysfunctions. This semiautomatic tumour detection system has a number of advantages over the existing systems given as follows: |
| 1. ATD is not only a method, but a multifunctional platform supporting real-time processing; |
| 2. It simultaneously detects organs as well as their pathology (tumours, stones and cysts) with increased accuracy; |
| 3. Processing timeis faster than the existing methods, as the main algorithms and additional controls run on the fly. The processing time for a 24 slices MRI dataset is about 1 min; |
| 4. 4) anoval mechanism for the seed pixel method avoids selection of irrelevant isolated pixels (implementing a top-down connectivity analysis between slices); |
| 5. The system achieves more accurate results for the recognition of kidneys compared with the existing methods by implementing additional controls. |
| The platform also supports storage of information in small anonymous xml-structured files (with a 500-kbytes typical file size for a complete dataset) to ensure fast transmission via networks to other clinicians. A typical size of a template file is about 2kbytes. |
METHODOLOGY |
| The method for creating and validating outputs of the ATD platform consists of three steps (see Fig. 1). |
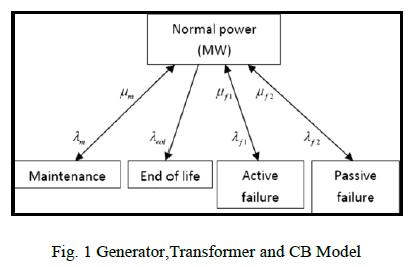 |
| A. “Three- Dimensional Golden Standard Models” The process calls for clinicians to annotate organs and pathologies in an abdominal MRI dataset in order to define Defining a golden standard model for the evaluation. For this process the manual segmentation panel integrates an advanced implementation of the region-growing semi automatic segmentation algorithm. Boundary refinement can be achieved by using a custom version of the pencil and eraser tools (keeping their neighbouring points connected) allowing for the expansion or shrinkage of the selected areas so as to add or remove any mistakenly selected or ignored areas. The integrated multilevel multifunctional annotation system facilitates the delineation of areas of interest in the selected image. Once the delineation process has been completed, the 3-D volume rendering option creates and enables the manipulation of the resulting realistic 3-D model of the organ structure and its potential abnormalities. The implementation uses visualization toolkit (VTK) [16], with visualization algorithms in a 3-D interactive process(Fig.2).Seven different views of the 3-D model are available along with corresponding metrics (see Fig.2 left side).They include MIP.(pH-sensitive Molecularly Imprinted Polymer), compositor amp, composite shaded amp, skin, bone, muscle and RGB composite. Newer volume rendering methods are in development [17], [18]. |
| B. Creating Templates with Rules and Parameters to Identify Specific Areas of Interest By experimenting with the parameters of the automatic regionoriented segmentation framework, a medical technician attempts to achieve a segmentation result closest to that of the clinician‘s using a region-oriented segmentation method [15]. A panel enables the interactive adjustment of the parameters of the framework in a single slide and the checking of the corresponding result in real time. Once the result is acceptable, the parameters and rules can be saved in a template with a name that corresponds to the area identified (e.g., right kidney) and can then be applied automatically to the entire image dataset. For highly complex images, the clinician has the option to define a working area where the Framework will be applied (see Fig. 3). Thevalidationpanel allowsanevaluationbycomparingtheresults forthetwomodelsforeveryslide.Thispanelshowsthetruepositives pixels which are the common pixels in the two methods (orange), the falsenegatives pixels which are presented only in the clinician‘s work (blue) and the false positives pixels which are presented only in the second image produced by the automatic method (green). |
| C.Using Existing Templates for Fully Automatic Identification of Specific Areas of Interest An end user can employ the existing templates to find a specific organ (e.g., left Kidney) and its potential abnormalities. The location of the organs can vary from one dataset to another; to identify the initial position of the organ, the engineer loads the template in any of the images including that organ and then clicks to verify the already selected organ from a list of objects. The system‘s functionality was tested on a dataset of 20 MRIs (522 images) acquired at a local regional hospital and with the following parameters: the MRI machine was a GE Medical Systems, running a scanning sequence of SE and a variant of SK. The slice thickness was 8 mm with a repetition time of 2000ms. Image frequency was 63.830539 MHz, magnetic field strength was 1.5 T, and spacing between slices was 10 mm. Avideodisplayingthe segmentation techniquesalongwithademonstration of the user interface and a working copy of the platform is available at: www.creteportal.gr/ATDSegmentation.zip. |
SEGMENTATION ALGARITHM |
| The segmentation framework is responsible for the training of the systems as well as the creation of the templates. The basic steps of this framework are as follows. |
| 1) A clinician opens an MRI dataset (i.e., abdominal). |
| 2) An edge-preserving anisotropic diffusion filter (Perona-Malik) removes the noise of the images [20]. |
| 3) The clinician selects the RoI, the organ, and the abnormalities and saves these choices. |
| `4) Later, a medical technician defines the working area thus reducing the amount of data to be processed. |
| 5) A modified version of a region-oriented segmentation algorithm is introduced to recognize the organs [15]. The histogram values are normalized to get better results: the user interactively defines the parameters to smooth the histogram (the up/down values to recognize more or less compact internal areas and thearea parameter to avoid selecting tiny objects during the organ‘s recognition process). |
| 6) The value of the seed pixel and the sensitivity are defined and used for the recognition of abnormalities. |
| 7) The medical technician records the best results and then stores the generated values into a template. |
| 8) Another user can now get a dataset of the same type (e.g., abdominal) and select a template to identify specific organs (e.g. Kidneys). The framework will automatically define the organ in the dataset and detect the potential abnormalities. |
| 9) Next, the key processes of the above steps are detailed. |
| A. Region Growing Semiautomatic Segmentation Algorithm |
| Starting from a seed pixel, the algorithm finds and demarcates structures as a mass of neighbouring pixels in image areas. It examines the intensity levels of the neighbouring pixels and recruits them as belonging to the structure being grown by spreading the selection outward starting from a predefined seed pixel whose gray-level intensity value, plus/minus a so-called tolerance value, forms the reference value for such recruitment of similar pixels [19]. The ATD platform integrates an enhanced version of this algorithm, to speed up the segmentation task: |
| 1) A slider allows the adjustment of the tolerance value interactively for the selection of the desired area in the image, just after the start point selection. |
| 2) An image mask (matrix) speeds up the segmentation process by ensuring that pixels already examined will not be checked again. |
| To solve the problem with the fuzzy transition zones at the boundaries of the designated areas of interest (e.g., kidney), advanced versions of the pencil and eraser correction tools are integrated in the platform (working with areas and not with pixels), to automatically keep the similarneighbouring points connected, providing clinicians with the option to swiftly refine their delineations. Processes of the above steps are detailed. |
| B. Region Oriented Segmentation Framework |
| In order to support automation of the diagnosis process, the proposed segmentation framework has 5 steps as follows. |
| 1) Pre-processing: The segmentation process starts with a pre-processingprocedure using an edge-preserving anisotropic diffusion filter[20], so that small image artefacts are smoothed (intraregionalsmoothing), while objects of interest such as edges are enhanced (inhibitinginter-regional smoothing). |
| 2) Rules Definition: Using a modified region-oriented segmentationalgorithm [15], the user can define a region of interest to minimizethe image information to be processed. Where there is no such definition of a working area where the organ is located, the automaticprocess would typically result in a larger number of areas being recognizedas objects of interest. Defined working areas are also stored intemplates. 3) Parameters Definition: In (1), the key parameters of the imageare defined: h(gv) is the histogram value for a specific gray-scale valuegv, Max is the maximum value of the histogram, and NormMax is thenormalized maximum value defined by the user. |
| h(gv) = int((float)h(gv) ∗ NormMax/Max). (1) |
| To identify the abnormalities located in a designated area, the histogramentries have to be compressed in such a way that the lowerentries of the histogram are emphasized. This is achieved by using (2). |
| h(gv) = 10_h(gv). (2) |
| `In this way, small peaks in the histogram corresponding to small areas in the image are emphasized, allowing the algorithm to detect them more accurately. A process of smoothing the histogram must also be applied to homogenize and successfully detect relatively large areas (organs) avoiding the insignificant local peaks (i.e., variations in luminance inside the organ‘s image area). The histogram is smoothed by applying the computational method whereby all the intensity values of the pixels in the image are recalculated by determining the average of a predefined number of neighbouring pixels of the histogram. This interpolation |
 |
| Width parameter is defined experimentally and interactively by the user. Higher width values are associated with larger areas being designated as part of the organ structure of interest, while low width values help the user pick out the smaller structures thus attempting to avoid missing any important parts of the image. The detection of the falling and rising histogram edges determines the number of valleys in thehistogram, thus inferring the threshold values (Lm): |
| Lm = a + down + [b − (a + down)]/2. (3) |
| In this equation, a represents a local maximum value, and b is the point where the histogram starts to rise again (ignoring any small peaks under a predefined limit value). These local minimum values (Lm) areused to segment objects and to create a label image. The closer the peak of this curve is to Lm, the more the delineated boundary moves inwards toward the centre of the kidney. The platform provides the clinician with the option to define the up and down parameters which are also stored in the templates in order to achieve the desired Lm values in complex images. Finally, applying any filter to an image introduces a certain amount of noise. This noise is removed with 3 × 3 erosion-dilation linear morphological filters. |
| 4) Connectivity Analysis: In order to create the final mark image which depicts the regions of interest, L-shape masks (based on a six connectedneighbourhood technique) are used [21] to compare the labelvalue of every pixel with the values of the neighbouring pixels (see Fig. 4). The pixels with the same values in the label image but different values in the initial image are included in the list of pixels to be considered later when identifying the number of the different objects in the image. |
RESULTS |
| Evaluation of the automatic segmentation framework, which is integrated in the ATD platform, is based on the following five criteria [22]. |
| 1) Accuracy, the proportion of true results (True Positives and True Negatives) in the total population of the results. |
| 2) Precision, the proportion of True Positives against all the positive results (True Positives and False Positives). |
| 3) Sensitivity, the ability of the system to identify positive results. It measures the proportion of positives which are correctly identified as such. |
| 4) Specificity, the ability of the system to identify negative results,meaning that it measures the proportion of negatives correctly identified as such. |
| 5) DSC, defines the similarity measure over two 3-D volume models (hereGSwhich is the Clinician‘s Golden Standard volume model versus MT which is the Medical Technician‘s volume model). |
| Although a medical technician is involved in this process, his/hersubjective opinion has very little effect on the overall process. Once the process is finished, the automatic results are compared with the output of the clinician‘s work. Therefore, any misguided selection performed by the medical technician will be cancelled out, since the main 3-D model of the findings has been initially defined by the clinician. |
| The average success rate for kidney detection is 97.2% and theirabnormalities 96.1% respectfully (see Table IV). As an example of the method the following results were obtained (see Fig. 5 and Tables I, II and III) from a random MRI dataset (SE00004): |
 |
| The above diagram depicts the integrated multilevel multifunctional annotation system. It facilitates in tracing the outline of the region of interest. Once the outline tracing process had been completed, the âÃâ¬Ãâ3-D volume rendering‘ option creates and enables the manipulation of resulting 3-D model of the organ structure and its potential abnormalities. |
 |
| Once the creation of Golden standard model was completed,the clinician selects the region of interest, organ and the abnormalities and saves these parameters in the form of template. Now the medical technicians experimentally adjust these rules and parameters for the integrated Automatic Recognition frame work to achieve results which are closest to those of clinicians, which is shown in the above figure.3.These parameters can later be used by non-experts to achieve the increased automation and identification process. |
 |
| In the above diagram the results are automatically compared with the output of the clinicians work. so if any misguided selection performed by the medical technician will be cancelled out. Since the main 3-D Model of the findings has been initially defined by the clinician. |
DISCUSSION |
| The proposed platform can achieve a highly accurate and fast identification of the kidneys, while being able to correctly identify any potential abnormalities (tumors, stones, and cysts) within the same platform. Overall, this platform supports the following features: |
| 1) A multi-functional easy-to-use annotation system. |
| 2) The ability for real-time detection of kidneys and their abnormalities (tumors, stones, and cysts). |
| 3) A multi functional 3-D visualization system providing real size measurements. |
| 4) Evaluation of the results based on five measurement methods including a 3-D volumetric method. |
| In order to deal with the possible inhomogeneity of images used, the platform performs the following steps. |
| `1) The Region of Interest is defined. |
| 2) The parameters used to accurately find the organ and its boundaries are defined: Width (for smoothing the histogram and get the correct boundaries of the organ), ascending and descending (for identifying local minimums), threshold (for removing the noise from the use of erosion—dilation morphological filters) and area (for initial identification of the organ ignoring tiny areas). |
| 3) The parameters to find the correct seeds for the pathology detection to feed to the region growing algorithm are also defined: Grayscale and a tolerance value for the range of values to represent a dysfunction, as well as a threshold value for the erosion filter used to change the sensitivity governing the discard of seeds that are not strong enough. By doing the above, the framework remains highly immune to datasets coming from different patients and even from different pathologies. Finally, the output files were made anonymous and were also of a small size and thus easier to upload. This would also enable the system to securely and efficiently transmit files, including images, to client Applications running on smartphones, so that clinicians and consultants could view the results on the move. |
CONCLUSION |
| This research presents a new MRI diagnosis-assistive platform that, after initial creation of a template, is capable of providing a more automatic 3-D identification of kidneys and their abnormalities (tumors, stones, and cysts). Two methods have been integrated to create 3-D volume models: The first provides clinicians with support (through a user interface) in order to rapidly identify and delineate areas of interest, with a 3-D view and have real metrics at hand specifying actual physical sizes of organ structures and any abnormal tissue regions. The second invokes a method of increased automation, to identify important areas based on templates that are initially created by a medical technician and later on used by a user with no particular prior knowledge of medical image segmentation. These templates allow the system to identify organ structures based on their features and look for any abnormalities. |
References |
|