ISSN ONLINE(2278-8875) PRINT (2320-3765)
ISSN ONLINE(2278-8875) PRINT (2320-3765)
S.Sivagowri1, Dr.M.C.Jobin Christ2
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Advanced Research in Electrical, Electronics and Instrumentation Engineering
Magnetic Resonance Imaging (MRI) can be used to detect lesions in the brains of Multiple Sclerosis (MS) patients and is imperative for diagnosing the disease and monitoring its progression. In practice, lesion load is often quantified by either manual or semi-automated segmentation of MRI, which is time-consuming, costly. We proposed model for automatic segmentation of multiple sclerosis lesions from brain MRI data. These techniques use a supervised classifier that is trained Support Vector Machine (SVM) to discriminate between the blocks in regions of MS lesions and the blocks in non-MS lesion regions mainly based on the textural features with aid of the other features. The main contribution of this set of frameworks is the use of textural features to detect MS lesions in a fully automated approach that does not rely on manually delineating the MS lesions. As a result, the sensitivity for detection of MS lesions was 81.5% with 2.9 false positives per slice based on a leave-one-candidate-out test, and the similarity index between MS regions determined by the proposed method and neuroradiologists. These results indicate the proposed method would be useful for assisting neuroradiologists in assessing the MS in clinical practice.
Keywords |
||||||||||||||
| Multiple sclerosis, Automatic lesion segmentation, Magnetic Resonance Imaging, Support Vector Machine. | ||||||||||||||
INTRODUCTION |
||||||||||||||
| Multiple Sclerosis (MS) is a chronic neurological disorder, which is caused by structural damages of axons and their myelin sheathes in the Central Nervous System (CNS). Depending on brain regions affected, MS could cause various central nervous system dysfunctions such as numbness or weakness of a limb, in coordination, vertigo or visual dysfunction. These clinical attacks are due to focal inflammation in the CNS directed against myelin, the insulation around nerve fibers (axons). The inflammation results in “MS Lesion”, which are characterized by demyelination, axon injury and axon conduction block. The progression of the MS lesions shows considerable variability and MS lesions portray temporal changes in shape, location, and area between patients and even for the same patient. Therefore, it is very important for radiologists to accurately detect MS lesions and follow-up the numbers, locations, and areas of MS lesions for diagnosis of each patient. | ||||||||||||||
| Magnetic Resonance Imaging (MRI) plays a vital role in quantifying brain lesion and other tissue for assessing the disease, understanding the underlying pathophysiology, and investigating the therapeutic efficacy in multiple sclerosis. Due to high resolution and good differentiation between soft tissues and other structures, MRI has superiority to the other imaging techniques for the studies of nervous system diseases. MRI has been known as the best paraclinical examination for MS which can reveal abnormalities in 95% of the patients [1]. Image segmentation or tissue classification is one of the critical steps for MR image analysis. The interpretation of MR images by a specialist is a difficult and time-consuming task, and result directly depends on the experience of the specialist. A reason for such a difficulty is related to the complexity and visually vague edges of anatomical borders [2]. Therefore, it is desirable to have an automatic segmentation method to provide an acceptable performance. | ||||||||||||||
| Several semi-automated and automated techniques have been developed for classifying lesions using images acquired in two-dimensional (2D) or three-dimensional (3D) mode with single or multiple MRI sequence(s). The multiple sequences include proton density-weighted (or PD), T1-weighted (or T1), T2-weighted (or T2), and Fluid Attenuated Inversion Recovery (FLAIR). Alfano et al. [3] proposed an automated approach based on relaxometric and geometric features for classification of MS lesions in 1.5 T 3-D MR images. Boudraa et al. [4] applied the FCM algorithm to 1.5 T two-dimensional (2-D) MR images for classifying normal and abnormal brain structures. Leemput et al. [5] proposed an automated method by using an intensity-based tissue classification and a stochastic model for detection of MS lesions in 1.5 T 3-D images. Zijdenbos et al. [6] developed an automated framework for the pipeline analysis of MS lesions in MRI data. Khayati et al. [7, 8] developed an automated method for segmentation of MS lesions in brain MR FLAIR images using an adaptive mixture method and a Markov random field model in 1.5 T 3-D MR images. However, the best of our knowledge most of the studies have focused on the segmentation accuracy of MS lesion. But have not reported the sensitivity and false positives, which are necessary for implementing a method in clinical situation. Therefore a sensitive and specific automated method to detect lesions in the brain is essential for the analysis of studies with a high numbers of MS patients. | ||||||||||||||
METHODOLOGY |
||||||||||||||
| In this paper we present an automatic MS lesion segmentation in brain MR images framework based on supervised voxel wise classification with an SVM classifier, which is a state-of- the-art machine-learning classifier. There are four main steps in our proposed systems shown in Fig.1. First, a pre-processing step includes registration of different MR modalities, intensity normalization, as well as in homogeneity correction. Second, step is used for the detection of initial MS lesions regions based on textural features. Third, the SVM model is used to perform the voxel-wise segmentation based on features. Finally, false positive voxels are further eliminated via post-processing techniques. | ||||||||||||||
A. IMAGE ACQUISITION |
||||||||||||||
| The multimodality images such as 2-D T1- weighted sequence, 2-D T2-weighted fast spin-echo sequence, 2-D FLAIR sequence from forty-nine slices of three cases (age range: 25 66,female:3)who took two studies with clinically diagnosed MS including 168 lesions were selected for this study. All images were acquired with a section thickness of 5mm, an intersection gap of 1mm, and a field of view of 22 cm, and the quantization levels in each pixel value were 16 bits. Fig. 2 (a)–(c) shows a T1-weighted image, a T2-weighted image, and FLAIR image, respectively, with an MS lesion indicated by a white solid line. The MS lesion has higher pixel values in the T2-weighted and FLAIR image, but lower pixel values in the T1-weighted image. | ||||||||||||||
B. PRE-PROCESSING |
||||||||||||||
| The following pre-processing steps are applied prior to the segmentation procedure: | ||||||||||||||
| Registration: MR sequences of the same patient are registered into the same space (patient space or stereotaxic space)).Registration might also be employed to align an atlas to the brain to provide some initial estimates of the brain tissues. For the MS Lesion Segmentation Challenge datasets, all datasets were rigidly registered to a common reference frame and re-sliced to isotropic voxel spacing, with resolution 512x512x512, using B-spline based interpolation. All the datasets were re-sliced to be in the same resolution conditions of MS Lesion Segmentation Challenge datasets to be tested by the models trained by MS Lesion Segmentation Challenge training data. Then they were registered to the MNI atlas using Automated Image Registration (AIR) software. | ||||||||||||||
| Brain extraction: The brain is extracted from the image, and the segmentation takes place only on the remaining brain voxels. At first, a head region was segmented by using an automated thresholding technique based on a linear discriminate analysis on the pixel-value histogram in the T1-weighted image, which usually had two main peaks corresponding to a head region and background, respectively. Next, a brain region was extracted by thresholding the pixel value, which was determined by using three times the standard deviation (SD) of the pixel-value histogram within the head region in the T1-weighted image for removing fat regions with high pixel values around the brain. | ||||||||||||||
| Intensity inhomogeneity (IIH) correction: The intensity of the same tissue can vary across the image due to inhomogeneity of the static or applied magnetic fields within the scanner. IIH correction methods reduce the smooth variation of intensity of the tissues to simplify subsequent segmentation. Contrast-brightness correction applied to maximize the intersection between the histogram of the training and segmentation datasets followed by using 3D anisotropic filter to eliminate empty histogram bins. | ||||||||||||||
| Noise reduction: The acquisition process can induce some noise in the image. MS lesions were enhanced by subtraction of a background image approximated by the first order polynomial in a brain region from the FLAIR image. Then, an unsharp masking filter was applied to the subtraction image for enhancement of boundaries of MS lesions. | ||||||||||||||
| Intensity normalization: Some segmentation methods require the intensity of the image to be similar to the intensity of the training images and thus depend on an intensity normalization step. Intensity normalization methods modify the intensity range of the target image and map them into a predefined intensity range. | ||||||||||||||
C. FEATURE EXTRACTION |
||||||||||||||
| In order to describe each square block of the MRI slice, a features vector of 39 features is calculated. The block features are classified into five categories: twenty four textural features, two position features, two co-registered intensities, three tissues priors and eight neighbouring blocks features. The textural features are calculated for the FLAIR sequence. Textural features include histogram-based features (mean and Variance), gradient-based features (gradient mean and gradient Variance), run length-based features (gray level non-uniformity, run length non-uniformity) and co occurrence matrix-based features (contrast, entropy and absolute value). Run length-based features are calculated 4 times for horizontal, vertical, 45 degrees and 135 degrees directions. Co-occurrence matrix-based features are calculated using a pixel distance d=1 and for the same angles as the run length-based features. The position features are the slice relative location with reference to the bottom slice and the radial Euclidean distance between the block’s top left pixel and the center of the slice normalized by dividing it by the longest diameter of the slice. The center and the longest diameter of the slice are parameters that are geometrically calculated in the pre-processing step. The other channels features include intensity means for the corresponding block in the T1and T2 channels. The atlas spacial prior probabilities features include the means of the priors extracted from the probabilistic atlas (White Matter, Gray Matter and CSF probabilities) for the block. The neighbouring blocks features are the difference between the mean intensity of the current block and the mean intensity of each of the eight neighbouring blocks in the same slice. | ||||||||||||||
D. SVM MODEL TRAINING AND SEGMENTATION |
||||||||||||||
1) Support Vector Machine (SVM): |
||||||||||||||
| Support Vector Machine (SVM) is a supervised learning algorithm, which has at its core a method for creating a predictor function from a set of training data where the function itself can be a binary, a multi-category, or even a general regression predictor. To accomplish this mathematical endeavour, SVMs find a hyper surface which attempts to split the positive and negative examples with the largest possible margin on all sides of the hyper plane. It uses a kernel function to transform data from input space into a high dimensional feature space in which it searches for a separating hyper plane. The radial basis function (RBF) kernel is selected to be the kernel of the SVM. This kernel nonlinearly maps samples into a higher dimensional space so it can handle the case when the relation between class labels and attributes is nonlinear. The library LIBSVM 2.9 [9] includes all the methods needed to do the implementation, training and prediction tasks of the SVM. It is incorporated in our method to handle all the SVM operations. | ||||||||||||||
2) Training: |
||||||||||||||
| The dataset of one or more subjects is used to generate the SVM training set. The slices of this training dataset are divided into n square blocks of size w × w pixels. SVM Training set T is composed of training entries ti (xi, yi) where xi is the feature vector of the block bi, yi is the class label of this block for i =1: n (number of blocks included in the training set). Our classification problem is binary, so yi is either 0 or 1. The training entry is said to be positive entry if yi is 1 and negative in the other case. For each slice of the training dataset, each group of connected pixels labelled manually as MS pixels forms a lesion region. Blocks involved in the positive training entries (TP) are generated by localizing all the lesion regions and for each of them, the smallest rectangle that encloses the lesion region is divided into non-overlapping square blocks of size w × w pixels. Each block bi of these blocks is labelled by yi = 1 if any of the w2 pixels inside this block is manually labelled as MS pixel. Any block that contains at least 1 MS pixel is defined in our method as MS block. Similarly, the blocks involved in the negative training entries (TN) are generated by localizing the non-back- ground pixels that are not manually labelled as MS pixels and dividing them into non-overlapping square blocks of size w × w pixels. Each block bi of these blocks is labelled by yi = 0. Feature vector xi is calculated for each block of both positive and negative training entries. The w2 MS pixels. This helps the SVM engine to learn the features of the blocks that either partially or completely contain MS pixels. | ||||||||||||||
| In our case, we used one subject dataset (only 10% of the subjects) which consists of thirty seven slices as the training dataset. Since the training set entries are as many as the number of blocks, the training set will be large enough (134173 training entries against 34 features with ratio 3946:1) to avoid the curse of dimensionality, which is the problem that the performances of the pattern classification systems could deteriorate if the ratio of the number of training data to that of features used for the classifier is relatively small. The training set entries were fed to the SVM engine to generate a MS classifier which is able to classify any square w × w block of a brain FLAIR MRI slice as MS block (y = 1) or non MS block (y = 0) based on its feature vector (x). | ||||||||||||||
3) Segmentation: |
||||||||||||||
| Each of the slices of the datasets to be segmented is divided into overlapping square blocks of size w × w pixels. The feature vector for each block is calculated. The trained SVM is used to predict the class labels for all the overlapping blocks. The block division is done in an overlapping manner to detect any possible MS blocks. For any block classified as MS block, assuming true positive classification, this does not mean that all pixels of the block should be classified as MS pixels because the SVM engine is trained to detect the blocks that contains MS pixels completely or partially. For each slice, all pixels are assigned an integer score. This score is initialized with a zero value. During segmentation, if any block is classified as MS block (y = 1), the scores of all pixels inside the block are incremented. As the blocks are overlapped, each pixel is part of w2 blocks as demonstrated in Fig .3 thus, the score will be any value from 0 to w2. Fig. 4 shows segmentation of sample slice from subject MS6 | ||||||||||||||
4) Post Processing: |
||||||||||||||
| The purpose of the post processing step is to improve and refine the performance of initial segmentation through dealing with different types of errors (false positives and false negatives). | ||||||||||||||
| Fig. 5 shows the initial segmentation of a sample slice from subject MS5 (Fig. 5(a)) and the colored evaluation of the segmentation in which the false negatives and positives are marked in green and red colors, respectively (Fig. 5(b)). | ||||||||||||||
| Errors in the initial segmentation of MS lesions can be classified as: | ||||||||||||||
| Type 1: False negatives resulting from not detecting MS lesion regions (labelled by 1 in Fig. 5(b)). | ||||||||||||||
| Type 2: False negatives resulting from incomplete MS lesion regions (labelled by 2 in Fig. 5(b)). | ||||||||||||||
| Type 3: False positives resulting from false MS lesion regions (labelled by 3 in Fig. 5(b)). | ||||||||||||||
| Type 4: False positives resulting from false portions of true MS lesion regions (labelled by 4 in Fig. 5(b)). | ||||||||||||||
5) Evaluation of the Proposed Method: |
||||||||||||||
| The performance of our method was evaluated by using a free-response receiver-operating characteristic (FROC) curve and overlap measure. The FROC curve was determined by changing the threshold value for the SVM output based on the leave one- candidate-out test. The overlap measure is calculated by the following equation, which denotes the degree of coincidence between the candidate region C obtained by our method and the grand truth region T by the manual method: | ||||||||||||||
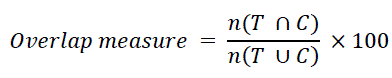 |
||||||||||||||
| where T is the grand truth region manually determined by two experienced neuroradiologists, C is the true positive region automatically determined by using our method, n(T ∪C) is the number of logical OR pixels between T and C, and n(T∩C) is the number of logical AND pixels regions between T and C. All candidate regions were classified into true positives and false positives based on the following criteria: if a “true” region included at least one pixel of a candidate region, the candidate region was classified as a true positive. Otherwise, the candidate region was considered as a false positive. | ||||||||||||||
RESULTS |
||||||||||||||
| At the initial identification step of MS candidates; our proposed method detected 93.5% of MS lesions with 101 false positives per slice. After applying the rule-based method, 47.5% of the false positives per slice were removed while keeping the sensitivity for detection of MS lesions. After applying the SVM, 84.3% of false positives per slice were excluded, whereas the sensitivity was decreased by 7.2%. Fig. 6 shows the effect of the SVM on classification of all candidate regions in FLAIR images with MS candidate regions. Some false positive regions similar to MS lesions were eliminated by the SVM. | ||||||||||||||
| For comparison with the other results, the average overlap measure (OM) of result in this study was converted into the similarity index =2OM/ (1+OM). This table does not include of Alfano et al. [1], because they did not explicitly state the similarity index in their papers. As a result the similarity index between MS regions determined by the proposed method and neuroradiologists was 0.77, which was highest among the past studies in Table I. | ||||||||||||||
| A higher the sensitivity and a lower number of false positive are essential for initial diagnosis and follow up of MS lesions. A more accurate segmentation of MS lesions is also necessary for diagnosing area change of MS lesions. MS lesion was identified and segmented well with an overlap measure of 81.8%. There were no false negative regions and only one false positive region in this slice. | ||||||||||||||
DISCUSSION |
||||||||||||||
| A novel method for MS lesions segmentation in FLAIR sequence of brain MR images has been developed. The segmentation process goes through three steps. The first step is the preprocessing which is done to improve the brightness and contrast of all the FLAIR slices of the subject dataset. The second step is the main processing which involves using a trained SVM to detect the initial MS lesions from the individual slices using feature vector which are mainly composed of textural features. The third step is the post processing that aims to improve and refine the performance of the initial segmentation generated through the SVM-based segmentation. In that regard, the post processing step addresses all possible types of errors in the MS lesion segmentation results of the second step in order to reduce the overall errors including both false positives and false negatives. The main processing classifier uses thirty four features in three categories. These categories of features are selected to have analogy with the features used nonintentionally by the expert in the task of manual labeling of MS areas. According to our observations, when the expert labels MS lesions in the FLAIR slice, the hyper intense areas are the potential areas to have the lesion. This is emulated in our technique by using the group of twenty four textural features. Candidate areas are filtered based on previous experience with the brain positions where most likely lesions occur; hence a group of two position based features is used. Besides, the expert takes into account the difference between the intensity of the lesion area and the neighboring areas intensities to take final decision and we emulate this by using the eight neighboring features. Although both textural features and neighboring features are based on intensity, no redundancy exists between them as the first group is used to aid the classifier in the detection of special pattern areas while the later is used to take into account the relation of the intensity of the area and the neighboring areas. The main processing classifier uses an SVM engine. SVM Parameters selection and training set balancing directly affect the classification performance. | ||||||||||||||
CONCLUSION |
||||||||||||||
| We have developed an automated method for detection of MS lesions in brain MR images using supervise classifier SVM. The main contributions of the presented method are using textural features without manual selection of ROI and the comprehensive post processing step that handles different types of errors in MS lesions segmentation that can be generalized to improve the performance of any other MS segmentation technique. Our preliminary results show that our proposed method may be useful for detection of MS lesions. As a result, although some modifications would be necessary for the detection of MS lesions, we believe that our method would be useful as one of the basic ideas for further development. | ||||||||||||||
Tables at a glance |
||||||||||||||
|
||||||||||||||
Figures at a glance |
||||||||||||||
|
||||||||||||||
References |
||||||||||||||
|