Department of Mathematical Sciences, Clemson University, Clemson, SC 29634-1907, USA
Received: 04/09/2015 Accepted: 09/11/2015 Published: 25/11/2015
Visit for more related articles at Research & Reviews: Journal of Statistics and Mathematical Sciences
In almost every field of study, studying the distribution of a random curve can give needed information. In the field of in vitro fertilization, particular interest is given to modeling the temperature curves of women across their menstrual cycle. Fitting the curve can lead to categorizing cycles as healthy or unhealthy and can help identify the most fertile days of a cycle.
Hierarchical model, Gibbs sampling, Simulation, Bayesian specification, Menstrual cycles.
In almost every field of study, studying the distribution of a random curve can give needed information. In the field of in vitro fertilization, particular interest is given to modeling the temperature curves of women across their menstrual cycle. Fitting the curve can lead to categorizing cycles as healthy or unhealthy and can help identify the most fertile days of a cycle. A healthy menstrual cycle consists of three phases, the follicular phase, ovulation, and the luteal phase. During the follicular phase, the basal body temperature (the lowest temperature reached during rest, referred to as bbt for the remainder of this paper) holds constant at a low plateau. During ovulation, the bbt increases until it hits the high plateau. The bbt then holds constant at this high plateau through the luteal phase. At the end of the cycle, the bbt drops back down to the low plateau before menstruation and the cycle repeats [1]. Figure 1 gives a few examples of a healthy menstrual cycle’s bbt levels [2]. Section 2 outlines the general hierarchical model, then gives a parametric modeling of the bbt curves. The end of Section 2 completes the Bayesian specification and specifies the hyperparameters needed for the prior distributions. Section 3 briefly describes how the Gibbs sampling algorithm works and then applies it to model. Section 4 gives a short description of the change-point problem and defines the stopping rule needed in order to identify the two change-points in the model. Section 5 details a simulation of the data and then applies the Gibbs sampling algorithm with intentionally skewed hyperparameters. The results of the simulation are discussed at the end of Section 5. Finally, Section 6 provides a short discussion of the model and conclusions.
Figure 1. Examples of bbt levels for healthy cycles[2].
First, we will define terms necessary to describing the model. A prior distribution of an unknown quantity X is the probability distribution that would express one’s certainty about X before the data is taken into account. Similarly, a posterior distribution of an unknown quantity X is the probability distribution of that quantity conditional on the observed data. A hyperparameter is parameter of a prior distribution that is not treated as random. For example, consider a random variable X that came from A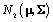 distribution, where Σ is known and we can use a
distribution, where Σ is known and we can use a 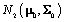 distribution to model μ. Then μ is a parameter,
distribution to model μ. Then μ is a parameter, 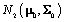 is a prior distribution, and μ0 and Σ0 are hyperparameters.
is a prior distribution, and μ0 and Σ0 are hyperparameters.
Let 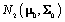 refer to woman i,jε {1,2,.....ni} refer to cycle j of woman i, and
refer to woman i,jε {1,2,.....ni} refer to cycle j of woman i, and 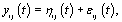 refer to day t in cycle j of woman i. These indices will retain these definitions throughout the remainder of the paper. We can establish the following model for bbt curves [3]:
refer to day t in cycle j of woman i. These indices will retain these definitions throughout the remainder of the paper. We can establish the following model for bbt curves [3]:
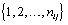
where εij is the measurement error and ηij is a smooth function from 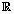 to
to  .Since cycles of the same woman are expected to be correlated, we can establish a prior distribution for the distribution of functions for woman
.Since cycles of the same woman are expected to be correlated, we can establish a prior distribution for the distribution of functions for woman 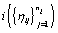 call it Gi . However, healthy menstrual cycles will follow a similar pattern among women, so we can establish a prior distribution for the collection of distributions for the different women
call it Gi . However, healthy menstrual cycles will follow a similar pattern among women, so we can establish a prior distribution for the collection of distributions for the different women 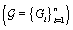 , call it P. Thus if we assume our error is normally distributed, we have the following model [3]:
, call it P. Thus if we assume our error is normally distributed, we have the following model [3]:
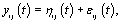
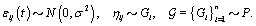
Basis representation
Now that we have our general model, there are a number of different representations that could be used for ηij. A common strategy is to consider a basis representation [3]:
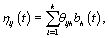
where 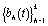 are pre-specified basis functions and
are pre-specified basis functions and 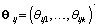 are cycle-specific basis coefficients. Since
are cycle-specific basis coefficients. Since 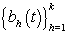 is prespecified, our major objective is to model the θijs. The approach we use is to give the θijs a hierarchical normal model [3]:
is prespecified, our major objective is to model the θijs. The approach we use is to give the θijs a hierarchical normal model [3]:
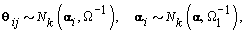
Where Nk(μ,Σ) is the multivariate normal distribution with mean vector μ and covariance matrix Σ. The woman-specific basis coefficients are given by αi, α gives the global mean basis coefficients, and Ω and Ω1 are the precision matrices for the within and between woman variability respectively. So to complete the model, we need to specify the basis functions 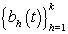 and establish the components of θij. We’ll do this in the next section when we consider a parametric hierarchical structure.
and establish the components of θij. We’ll do this in the next section when we consider a parametric hierarchical structure.
Parametric Hierarchical Model
As we discussed in Section 1, bbt levels of a healthy cycle follow a biphasic pattern where the bbt levels start at the low plateau in the follicular phase then quickly rise during ovulation to the high plateau in the luteal phase. Thus we can model ηij (t) as follows [3]:
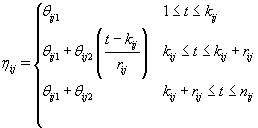
where 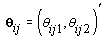 where θij1 and θij2 indicate the temperature during the follicular phase and the increase in bbt during ovulation respectively for cycle j of woman i. We can further define to be the last day of the follicular phase and rij to be the number of days the bbt rises during ovulation (Figure 2). Thus, if we consider yij (t) as Yij a vector of nij bbt values, we can represent our hierarchical model in matrix form [3]:
where θij1 and θij2 indicate the temperature during the follicular phase and the increase in bbt during ovulation respectively for cycle j of woman i. We can further define to be the last day of the follicular phase and rij to be the number of days the bbt rises during ovulation (Figure 2). Thus, if we consider yij (t) as Yij a vector of nij bbt values, we can represent our hierarchical model in matrix form [3]:
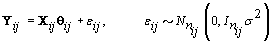
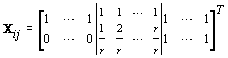
Thus Xij (t), the tth row of X, corresponds to tth day of cycle j of woman i. So we have defined our pre-specified basis function 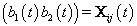 and our cycle-specific basis coefficients
and our cycle-specific basis coefficients 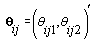 .
.
Bayesian Specification
Now we have within (Ω) and between (Ω1) woman variability, the last day of the follicular phase (kij), the number of days during ovulation (rij), the global mean (α), and measurement error variance (σ2) left to model. We’ll use the following equations to model the within and between woman variability:
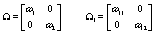
We complete the Bayesian specification with the following priors [3]:
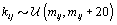
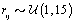
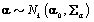
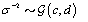
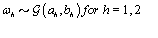
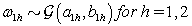
where 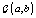 is the gamma distribution with shape parameter a and inverse scale parameter b,
is the gamma distribution with shape parameter a and inverse scale parameter b, 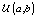 is the discrete uniform distribution between a and b, and mij is the first day after menstruation. Thus, our pre-specified hyperparameters are α0, Σα, c, d, ah, bh, ah1, and bh2 (for h = 1, 2).A summary of the parameters, hyperparameters, their descriptions, and distributions is given in the appendix.
is the discrete uniform distribution between a and b, and mij is the first day after menstruation. Thus, our pre-specified hyperparameters are α0, Σα, c, d, ah, bh, ah1, and bh2 (for h = 1, 2).A summary of the parameters, hyperparameters, their descriptions, and distributions is given in the appendix.
Our goal is to find the parameters of our model given a set of observations. Since finding the joint distribution is a complicated process, we need to use a different method. A common method to do this is to use a Monte Carlo Markov Chain as an iterative procedure. The specific type of Monte Carlo Markov Chain that we’ll use is called a Gibbs sampler.
Suppose we have random variables X1, X 2,…, X n.
1. Initialization: Let 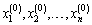 be given some initial value.
be given some initial value.
2. Iteration: For 1 ≤ I≤ K, sample x (i)j from the conditional distribution of Xj given 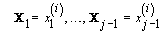 and
and 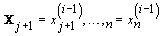 for 1 ≤ j ≤ n
for 1 ≤ j ≤ n
This Gibbs sampling procedure is a Markov chain, where the stationary distribution is joint distribution of X1, X 2,…, X n.Thus, for large k, the sample 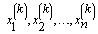 approximates the joint distribution of X1, X 2,…, X n. .A blocked Gibbs sampler uses the same steps, except it groups some of the random variables together so that the variables in a block are sampled from their joint distribution conditioned on all other variabes.
approximates the joint distribution of X1, X 2,…, X n. .A blocked Gibbs sampler uses the same steps, except it groups some of the random variables together so that the variables in a block are sampled from their joint distribution conditioned on all other variabes.
Gibbs Sampling Algorithm
Here is the blocked Gibbs sampling algorithm for our model after we’ve given the parameters some initial value [2]:
1. Cycle-specific coefficients: 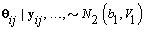
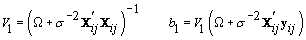
2. Women-specific means: 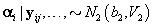
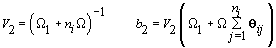
3. Global mean: 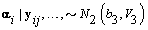
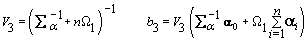
4. Components of Ω( h = 1,2): 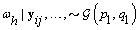
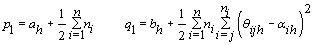
5. Components of 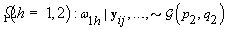
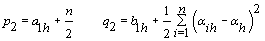
6. Error variance: 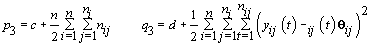
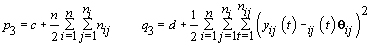
7. Last day of follicular phase: Use the change-point stopping rule for kij (see Section 4).
8. Ovulation period: Use the change-point stopping rule for rij (see Section 4).
So if we use the blocked Gibbs sampling steps to update the unknown parameters from their conditional distributions, then after a large number of iterations, we’ll be sampling from the joint distribution of all the unknown parameters.
Now we need to consider kij and rij since these are specific points (called change-points) in the bbt curve where the shape of the curve changes. The process for finding a change-point P is intuitive. Given a set of observations 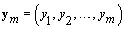 .we’ll find the probability that p ≤ m given ym . We want this probability to be higher than some high threshold Q. Thus we have a “stopping rule" [4]:
.we’ll find the probability that p ≤ m given ym . We want this probability to be higher than some high threshold Q. Thus we have a “stopping rule" [4]:
Terminate at m if and only if 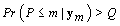
Starting with m = 1, the first time that the probability is greater than Q is our change-point P.
First we wish to find kij assuming that the other parameters are known. Let Xk be the first m rows of the matrix Xij established above but with kij = k. Thus 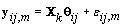 . Let {πk} be the prior distribution for kij,π(.) be the density function of θij, and f(.) be the density function of the data yij,m. We now need to find the posterior distribution of kij in order to find
. Let {πk} be the prior distribution for kij,π(.) be the density function of θij, and f(.) be the density function of the data yij,m. We now need to find the posterior distribution of kij in order to find 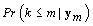 for our stopping rule.
for our stopping rule.
Let 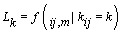 be the change-point likelihood. Then implementing Bayes’ theorem, we get [4]
be the change-point likelihood. Then implementing Bayes’ theorem, we get [4]
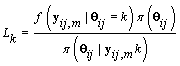
Since Lk is not a function θij we may replace θij with 0. Also 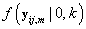 does not depend on k, so Lk is proportional in k to
does not depend on k, so Lk is proportional in k to 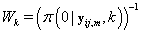
The posterior distribution of θij is N2(μk,Vk) where
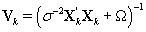
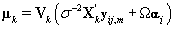
Note that this is the same posterior distribution found for θij found in Section 3, but with the altered matrix XK . Using this posterior distribution for θij , we find that
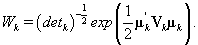
Since we need to find 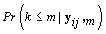 , we’ll first find
, we’ll first find 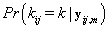 .From applying Bayes’ theorem to
.From applying Bayes’ theorem to 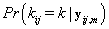 , we get
, we get 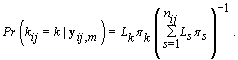
Since LK is proportional in k to WK, then we can substitute Wk in for LK , resulting in
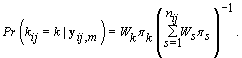
These probabilities are easy to compute as the data becomes available. When k ≥ m,
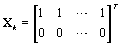
So XK does not depend on k (and hence WK = WM). Thus, our stopping rule is to terminate at m if and only if
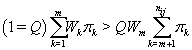
Once we hit our stopping rule [4], let kij =m.
Finding rij
We can apply the same approach to finding rij by using a transformation to examine the data “backwards", i.e. consider the last day first, and proceed through the cycle in reverse order. So our new reversed data is
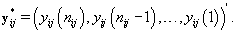
Let 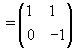 be our transformation matrix. Then
be our transformation matrix. Then
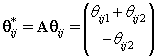
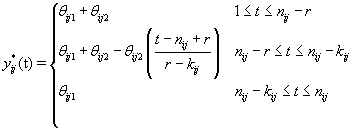
Thus if we let Xr* correspond to the first m rows of the above system, then
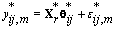
This transformation lets us use the same process that we used to find kij. Let {πr } be the prior distribution for r (the second changepoint). The new posterior distribution for θ*ij is N2(μr,Vr), where
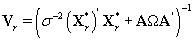
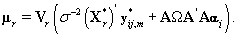
Note that since 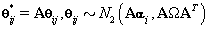 thus giving a similar posterior distribution found in Section 3 for θij. Using this posterior distribution for *ijwe find that
thus giving a similar posterior distribution found in Section 3 for θij. Using this posterior distribution for *ijwe find that
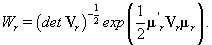
Thus, our stopping rule is to terminate at m if and only if
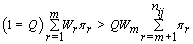
Once we hit our stopping rule, r =nij-m (since we transformed our data). Since rij is the number of days between kij and r, then rij = r –kij
Now that the method has been described, we’ll introduce a simulation in order to illustrate the method. The simulated data consisted of 20 women, each with 10 cycles, so we had a total of 200 cycles. Each cycle had a length of 30 days [5]. We arbitrarily assigned the global mean and covariance matrices to be
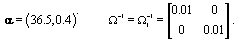
Each woman specific mean was generated as per the model discussed in Section 2, so 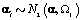 We then generated kij and rij for each cycle according to
We then generated kij and rij for each cycle according to 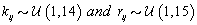 .Finally, we generated the cycle specific coefficients according to
.Finally, we generated the cycle specific coefficients according to 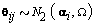 and the measurement error terms according to
and the measurement error terms according to 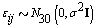 where σ 2= 0.01. Now that we generated all the parameters needed for our model, we generated the bbt values according to
where σ 2= 0.01. Now that we generated all the parameters needed for our model, we generated the bbt values according to
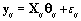
In order to run the algorithm, we need to specify the hyperparameters (our initial guesses). In order to account for error, we intentionally perturb the hyperparameters away from the actual parameters established in Data simulation Section So we set our hyperparameters to be
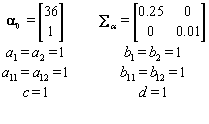
These hyperparamters are used in the Gibbs sampling steps outlined in Change-point probleam section. But before we can implement the Gibbs sampling algorithm, we need to initialize our unknowns from their prior distributions using the hyperparameters, as seen below:
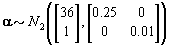
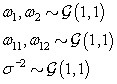
We also need to initialize the αis from their prior distributions since the first Gibbs sampling step is finding the posterior distribution of θij conditional on αi . So we’ll generate αi values from the prior distribution
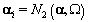
The final step for initialization was to set the kij and rij values all to a fixed value of 7.
Now that initialization is completed, we can run the algorithm. We ran the algorithm for a total of 6,000 iterations with a 1,000 iteration burn-in. To make sure that the parameters were converging, we performed traceplots of a number of the variables. Figures 3 and 4 show the traceplots for α11 and α12 respectively.
Tables 1 and 2 summarize the results from estimating the parameters in our model. The cycle specific parameters (kij, rij,θij1 and θij2) are summaries over all cycles, while the women specific parameters (αi1 and αi2) are summaries over all women. As evidenced by the tables, the distribution of these parameters was well estimated. Figures 5 and 6 show graphs of the true and estimated distributions for two example cycles.
So, we can see that the blocked Gibbs sampling algorithm was an accurate method for estimating the various parameters of our hierarchical model. Most importantly, the change-point analysis gave accurate estimates for the last day of the follicular stage and the number of days that the temperature rises during ovulation (Appendix). These two figures are critical for determining the most fertile period of the menstrual cycle. A natural extension of this topic would be to model unhealthy cycles using a nonparametric approach. Combining the parametric and nonparametric models would allow for broader analysis and more unusual data sets.