Keywords
|
| Object Counting, Image Processing, Image Segmentation. |
INTRODUCTION
|
| Object counting is a very common task performed in different industries. Figuring out how many objects in an image is required in image analysis. Object counting is used to get certain number of elements from images. These elements act as a source of information for quantitative analysis, motion tracking and qualitative analysis. |
| The conventional method for object counting is manual, time consuming and in non-automatic form. Continuous counting leads to eye fatigue and affects the accuracy of results. However, the process of counting objects is not always straightforward or trivial, even performed manually. Most counting methods have peculiarities that make them tricky to tackle. For example, the objects may occur in large number and overlapped making counting tricky and tedious that in turn leads to error. Manual method must be replaced by computer vision as the results of this method are erroneous and time consuming [1]. Automatic counting of objects is a subject that has received significant attention in last few years with objects as varied as cells [2], RBCs [3], fish [4], eggs [9] etc. |
| Because automatic counting is objective, reliable and reproducible, comparison of cell number between specimens is considerably more accurate with automatic programs than with manual counting. While a user normally gets a different result in each measurement when counting manually, automatic programs obtain consistently a unique value. Thus, although some cells may be missed, since the same criterion is applied in all the stacks, there is no bias or error. Consistent and objective criteria are used to compare multiple genotypes and samples of unlimited size. |
| Cell counting is very important and useful for medical diagnosis and biological research. Counting microorganisms and colonies is one of the most basic activities in health tests, food quality control, agriculture analysis etc [2]. Blood count is one of the most commonly performed blood test in medicine. It is required to detect as well as to follow disease treatment [3]. In marine science research, fish population estimation and fish species classification is important for the assessment of fish abundance, distribution and diversity in marine environments [4]. Object counting is also needed in some other research fields where objects cannot be segregated by naked eye and the factors ‘time’ and ‘accuracy’ matter |
| It becomes challenging when different objects are not easily distinguishable, vary in size and surrounded by noisy background. It is important to notice the variety of objects being counted as the accuracy of development algorithm is dependent on the same. |
| Currently, there is lot of research is being made on object counting. This paper presents literature review of few of the methodologies of object counting. |
II.FRAMEWORK
|
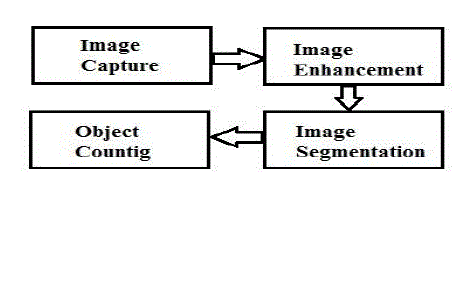
| The general method for object counting follows the following framework: |
| 1. Image Acquisition: This step intends to capture image through camera. The quality of image depends on camera parameters, lighting conditions, size of objects and distance from which image is taken. For better results, cameras with higher resolution are preferred |
| 2. Image Enhancement: Objective of image enhancement is to process image so that resulting image is more suitable than original image for specific application. During this process, one or more attributes of image are modified. The choice of attributes and the way in which image should be modified depend on specific application. These include basic gray level transformations, histogram modification, average and median filtering etc |
| 3. Image Segmentation: Segmentation is used to partition an image into distinct regions containing each pixel with similar attributes. The result of segmentation is set of segments that collectively cover entire image or set of contours extracted from image. It can be region based segmentation or data clustering or edge based segmentation. |
| 4. Object Counting: Object counting is done get number of segmented areas. Some of the methods of object counting are blob analysis, connected components analysis, statistical area measurements etc. |
III.LITERATURE REVIEW
|
| 1. Xiaomin Guo and Feihong Yu introduced a method of automatic cell counting based on microscopic images [2]. Histogram information is used to calculate adjustable lower and upper threshold value. This value is used for segmentation of objects and background. Effect of Floodfill method fills the objects region. It is used to mark or separate regions in an image. A blob is an area of touching pixels with the same logical state. All pixels in an image that belong to a blob are in a foreground state. All other pixels are in a background state. Blob analysis is used to detect blobs in an image and make selected measurements of those blobs. Blob analysis consists of a series of processing operations and analysis functions that produce information about any 2D shape in an image. If size of a blob is beyond the upper threshold of area, the blob will be segmented by K-means clustering algorithm. By calculating the number of cells contained in each blob obtains the total number of cells in whole image. The result shows that maximum relative error is 1.33%, minimum relative error is 0% and the average relative error is 0.46%. |
| 2. Venkatalakshmi. B et al. presented a method for automatic red blood cell counting using hough transform [3]. The algorithm for estimating the red blood cells consists of five major steps: input image acquisition, pre-processing, segmentation, feature extraction and counting. In pre-processing step, original blood smear is converted into HSV image. As Saturation image clearly shows the bright components, it is further used for analysis. First step of segmentation is to find out lower and upper threshold from histogram information. Saturation image is then divided into two binary images based on this information. Morphological area closing is applied to lower pixel value image and morphological dilation and area closing is applied to higher pixel value image. Morphological XOR operation is applied to two binary images and circular hough transform is applied to extract RBCs. |
| 3. J.N. Fabic et al. described an efficient method for fish detection, counting and species classification from underwater video sequences using blob counting and shape analysis [4]. The proposed system is consists of four major steps: Pre-processing, Contour detection, Blob Counting and Species Identification. Preprocessing is done for cleaning the background by eliminating unwanted objects. It involves Coral Blackening Procedure to blacken out corals using color histogram, Inward-Outer Block Erasure Algorithm to distinguish between fish and water and Edge Cleaning Algorithms for clearly defining edges. Contour Detection utilizes the Canny edge detection to detect fish contours and fill up spaces to allow blob counting. The blob detector is based on Laplacian of Gaussian (LoG). Connected components algorithm is used to label connected regions in binary images and subsets can be uniquely extracted. These results are used for counting, filtering or tracking. Species identification is done with the help of image moment features of the blob. From results, it is observed that the tolerance is less than 10 %. |
| 4. Haider Adnan Khan et al. presented a framework for cell segmentation and counting by detection of cell centroids in microscopic images [5]. Preprocessing is done with Contrast-Limited Adaptive Histogram Equalization to get enhanced image. Next, cells are separated from background using global thresholding. Then, distance transform of binary image is computed which converts binary image into distance map indicating distance of every cell pixel from its nearest background pixel. In order to perform template matching, the template image is generated from the distance transform of circular disk. Distance map is used to identify the cell centroids. The template matching is done using normalized cross-correlation between template and distance map. Finally, the similarity matrix is complemented and all background pixels are set to -∞. The watershed transform is then applied on this complemented similarity matrix. This splits the similarity matrix into separate disjoint regions. Each region is labeled and counted to get the count. The experimental results show excellent accuracy of 92 % for cell counting even at very high 60 % probability. |
| 5. Watcharin et al. proposed an algorithm to count blood cells in urine sediment using ANN and hough transform [6]. First step of algorithm is the segmentation between background and blood cells by using feedforward backpropagation algorithm. For training neural network, the input is Hue, Saturation, Value and standard deviation. After deriving output from feedforward backpropagation, salt and pepper noise is eliminated by using morphological opening and closing method. Last step is blood cell counting using circular hough transform. Experimental results show the average percentage of error of RBCs and WBCs detection 5.28 and 8.35 respectively. |
| 6. J. G. A. Barbedo presented a method for counting of microorganisms that use a series of morphological operations to create a representation in which objects of interest are easily isolated and counted [7]. First step of this method is RGB to gray conversion. After that, two-dimensional median filter is applied, in order to eliminate noise and other artifacts. Ideal size of the neighborhood over which filter should be applied depends on three main factors: size of objects of interest, size of spurious artifacts and resolution of the image. The program has two approaches for deciding neighborhood. In the first approach, user enters estimate of diameter of objects and artifacts. In the second approach, estimation using multiple counts is done. Then, contrast is adjusted in such a way the brightest pixel assumes the full-scale value 255 and darkest pixel equal to zero. In following, the algorithm verifies if the background is brighter or darker than the objects. If the background is brighter, a complement operation is performed. The image is then submitted to top-hat morphological filtering. Image is binarized with threshold in 128. After that object counting becomes trivial. By observing results, it can be seen that, except for the case of merged objects, the method identifies the objects correctly in more than 90 % of the cases, and the number of false positives is always low. The overall deviation was 8 %; such a number falls to 2.5 % if the images with merged objects are not taken into account. |
| 7. Marjan Ramin et al. used image analysis technique for counting number of cells in Immunocytochemical (ICC) images [8]. The proposed system contains four major steps: Pre-processing, Classification, Separating Bound Nucleus and Cell Counting. Pre-processing consists of removal of random noise by smoothening spatial filter. Morphological open operator is utilized to eliminate images’ background. Banding noise is removed by subtracting median of the red channel from all channels. In order to separate nucleus from antigens, nearest neighbor classification method with Euclidean distance metric is used in L*a*b color space. The bound nucleus is separated by local thresholding algorithm. For this purpose, statistical analysis is done and optimal threshold is found with the help of genetic algorithm. Finally, cell counting is done by tracing the boundaries. From the results, the Error Ratio and Standard Deviation of the proposed method are 6.75% and 6.39% respectively. |
| 8. Carlos A. B. Mello et al. presented two methods for mosquito eggs counting. These methods are based on a different color model [9]. In the first method, RGB image is converted into HSL color model (Hue, saturation, Lightness). From these three components, the hue image is extracted as it contains information about color tone. Huang thresholding algorithm is applied to the hue image for binarization. A connected components algorithm is used to label the connected regions of the image. Filtering is done using morphological opening operation with structuring element defined in the form of egg. At the last step, it is considered that egg occupies area of 170 pixels. The number of eggs is calculated by dividing the total amount of white pixels by this average area. The second method is based on converting RGB sub-image to YIQ one. From these components, I band is segmented in two ways: by using limiarization with fix threshold of 200 and by binarization using k-means clustering method. For performing egg counting in this method, it is considered that the average size of mosquito egg is 220 pixels. |
IV.APPLICATIONS
|
| Object counting using image processing has huge applications where automation is to be introduced and time of counting is to be reduced. Some of the main applications of object counting in industrial systems are packaging, quality control, and so on. It is helpful in the research areas where objects are of very small size. Object counting algorithm can be also used to track and identify objects. The present methods can be extended to have counting system based on userselected attributes. |
V. CONCLUSION
|
| Image processing techniques are helpful for object counting and reduce the time of counting effectively. Proper recognition of the object is important for object counting. The accuracy of the algorithm depends on camera used, size of objects, whether or not objects touching and illumination conditions. |
Figures at a glance
|
 |
| Figure 1 |
|
| |
References
|
- Jayme Garcia ArnalBarbedo, “Automatic Object Counting in Neubaur Chamber,” Proc. of XXXI Brazilian Telecommunications Symposium,Fortaleza, Dec. 2013.
- Xiaomin Guo and Feihong Yu, “A Method of Automatic Cell Counting Based on Microscopic Image,” 5th International Conference onIntelligent Human-Machine Systems and Cybernetics, Vol. 1, Aug. 2013. pp. 293-296.
- Venkatalaksmi. B and Thilagavathi. K, “Automatic Red Blood Cell Counting Using Hough Transform,” Proc. of 2013 IEEE Conference onInformation and Communication Technology, Apr. 2013, pp. 267-271.
- J.N. Fabic, I.E. Turla, J.A. Capacillo, L.T. David and P.C. Naval, Jr, “Fish Population Estimation and Species Classification from UnderwaterVideo Sequences using Blob Counting and Shape Analysis,” 2013 International Underwater Technology Symposium (UT), Mar. 2013. pp. 1-6.
- Haider Adnan Khan and GolamMorshedMaruf, “Counting Clustered Cells using Distance Mapping,” 2013 International Conference onInformatics, Electronics and Vision (ICIEV), May 2013. pp. 1-6.
- WatcharinTangsuksant, ChuchartPintavirooj, SomchartTaertulakarn, SomsriDaochai, “Development Algorithm to Count Blood Cells inUrine Sediment using ANN and Hough Transform,” The 2013 Biomedical Engineering International conference, Oct. 2013. pp. 1-4
- Jayme Garcia ArnalBarbedo, “Method for Counting Microorganisms and Colonies in Microscopic Images,” 12th Int. Conf. Computer Scienceand Its Applications, June 2012. pp. 84-87.
- Ramin M., Ahmadvand, P., Sepas-Moghaddam, A.Dehshibi, M.M., “Counting Number of Cells in Immunocytochemical Images usingGenetic Algorithm,” 12th International Conference on Hybrid Intelligent Systems, Dec. 2012. pp. 185-190.
- Carlos A. B. Mello, Wellington P. dos Santos, Marco A. B. Rodrigues, Ana LciaB.Candeias, Cristine M. G. Gusmao ,“Image Segmentation ofOvitraps for Automatic Counting of AedesAegypti Eggs,” 30th Annual International IEEE EMBS Conf. Vancouver, British Columbia,Canada, Aug. 2008. pp. 3103-3106.
- G. Gusmao, Saulo C. S. Machado, Marco A. B. Rodrigues, “A new Algorithm for Segmenting and Counting AedesAegypti Eggs inOvitraps,” 31st Annual International Conference IEEE EMBS Minneapolis, Minnesota, USA, Sep. 2009 pp.6714-6717.
- Y. H. Toh, T.M. Ng, B.K. Liew, “Automated Fish Counting using Image Processing,” International Conference on Computational Intelligenceand Software Engineering, CiSE, Dec. 2009. pp. 1-5
- Yan Wenzhong, “A Counting Algorithm for Overlapped chromosomes,” The 2nd International Conference on Bioinformatics and BiomedicalEngineering, June 2009 pp. 1-3.
- ChomtipPornpanomchai, FuangchatStheitsthienchaiSorawatRattanachuen, “Object Detection and Counting System”, 2008 Congress onImage and Signal Processing, May 2008. pp. 61-65.
- Qingmin LIAO, Kacem CHEHDI , Xinggang LIN, Yujin ZHANG, “Identification of Pelagic Eggs by Image Analysis,” 3rd InternationalConference on Signal Processing, Oct. 1996. Vol.2. pp. 855-858
|