ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Dr. B. Kwakye-Awuah1,*, Dr. A, Mrozik2 , Prof. Z. Piotrowska-Seget2, I. Nkrumah1 Prof. C. Williams3, Dr. I. Radecka3
|
| Corresponding Author: Dr. B. Kwakye-Awuah,Lecturer, Department of Physics (Materials Science group), KNUST, Kumasi, Ghana, Tel. +233(0)545207159, Email: bkwakye-wuah.cos@knust.edu.gh |
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
The kinetics of the release profile of silver ions from zeolite X framework and its effect on fatty acid composition of two gram-negative: Escherichia coli K12 W-T and Pseudomonas aeruginosa NCIMB 8295 and one gram-positive Staphylococcus aureus NCIMB 6571. Silver ions were released from the zeolite framework in an anomalous manner with time. The fatty acid composition was significantly altered when they were exposed to silver-loaded zeolite X. Interaction of silver ions with fatty acid of bacterial cells is likely to affect the survival of the cell
Keywords |
| Silver-loaded zeolite X, fatty acid, bacteria, silver ions |
I. INTRODUCTION |
| Silver ions have been reported to affect many different functions of microbial cells and non-selective in its antimicrobial action [26] – [27], [33]. Hence, silver ions have antimicrobial activity against a broad spectrum of microorganisms [9] – [11]. Since the ionic species carries a strong positive charge it has high affinity for negatively charged groups of biological molecules [24]. These molecules contain groups such as sulfhydryl, carboxyl, phosphates and esters commonly found on macromolecular structures distributed throughout microbial cells. Silver ions have also been reported to effectively inactivate many functions such as cell wall synthesis, membrane transport, nucleic acids such as RNA and DNA synthesis and translation, and protein folding and function (Russell and Hugo, 1994; Sondi and Salopek-Sondi). The binding of the Ag+ was found to alter the molecular structure of the macromolecule, rendering it worthless to the cell [3]. The exact mechanism of antimicrobial effect of silver on the bacterial cells is not well understood [9, 32, 33]. For instance whilst Feng et al [9] reported that the cytoplasmic membrane shrank and detached from the cell wall on exposure to silver ions, observations by Yamanaka et al [37] indicated that bactericidal actions of the silver ions are caused primarily by its interaction with the cytoplasm in the interior of the cell. Furthermore, the cytoplasmic membrane has been found to be a key target for investigation into bacterial stress response [18]. According to Mykytczuk et al [18], heavy metal cations interact with lipid membrane properties of bacterial cells. Such interaction causes alters membrane fluidity and phase characteristics as well as overall membrane integrity. This study was undertaken to determine whether there exist a relationship between the fatty acid composition of bacterial cells and their susceptibility to silver ions. Cultures of two gram-negative bacteria; Escherichia coli (E. coli) K12 W-T and Pseudomonas aeruginosa (P. aeruginosa) NCIMB 8295 and one gram-positive bacteria Staphylococcus aureus (S. aureus) NCIMB 6571 in their mid-exponential phase were harvested and challenged with silver-loaded zeolite X. Zeolites are crystalline aluminosilicate inorganic materials whose framework structure consist of cavities or pores that are occupied by cations or water molecules [12, 13, 19]. The zeolite serves as an inorganic reservoir for the silver ions and releases it in a slow and continuous manner when solvated. Changes in cellular fatty acid composition were analysed as well as variations in the saturated/unsaturated components. In addition, the release profile of silver ions from the zeolite X framework was investigated. |
II. MATERIALS AND METHOD |
| An easy way to comply with the conference paper formatting requirements is to use this document as a template and simply type your text into it. |
| A. Reagents, bacteria, media, zeolite X and silver-zeolite X |
| Sodium hydroxide and sodium aluminate were purchased from Aldrich Chemicals, UK. Sodium silicate was purchased from Fischer Chemicals, UK. (TSA) and Tryptone soya broth (TSB) were obtained from Lab M, UK. De-ionized water used in this investigation was supplied by University of Wolverhampton. Zeolite X (batch composition: NaAlO2:4SiO2:16NaOH:325H2O) without silver and silver-loaded zeolite X were synthesized and characterize by Kwakye-Awuah (2008). Bacterial strains; E. coli K12 W-T, S. aureus NCIMB 6571 and P. aeruginosa NCIMB 8295 used in this study were obtained from the University of Wolverhampton culture collection. The stock cultures were freeze-dried and kept at –20 oC overnight. Before use, cultures were resuscitated and grown on Tryptone soya agar (TSA) overnight at 37 oC. |
| B. Extraction and analysis |
| Before the extraction process, extraction reagents (saponification, methylation, extraction solvent and base wash) were first prepared. Saponification reagent was prepared by first diluting 150 ml of methanol (HPLC grade; Aldrich Chemicals, UK) with 150 ml of de-ionized water and adding 45 g of sodium hydroxide. The resulting solution was stirred until the sodium hydroxide was completely dissolved to obtain a methanolic solution. To prepare the methylation reagent, 325 ml of 6.0 N HCl was added to ethanol with constant stirring until a uniform solution was obtained. Extraction solvent was prepared by adding 200 ml of methyl tert-butyl ether (MTBE) to 200 ml hexane (HPLC grade) with uniform stirring for about 5 minutes to ensure a homogeneous solution. Finally base wash reagent was prepared by dissolving 10.8 g of sodium hydroxide in 900 ml de-ionized water. A single bacterial colony of each microorganism was used to inoculate a 100 ml starter culture, which were grown aerobically in TSB (pH 7.2) overnight at 37 oC in a rotary shaker (150 rev min-1) (Bellantone et al., 2000; Kwakye-Awuah et al 2008a). After overnight |
 rotary shaker (150 rev min-1). In the first stage, 0.2 g (1.0 gl-1) of silver zeolite was added immediately after inoculation. Samples were taking every 15 minutes for two hours. The concentrations of silver ions over each time period were determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES). In the second stage the inoculated media were incubated at 37 oC in a rotary shaker (150 rev min-1) for two hours before exposing them to 0.2 g of silver-loaded zeolite X (1.0 gl-1). Zeolite X without silver ions was used as control. Cells in the late exponential phase were harvested by centrifugation (6900 × g) at 15 oC for 10 minutes and transferred into culture tubes. The extraction was carried out as follows (Sasser 1990): One millilitre of the saponification reagent was added to the cells in the culture tubes (two replicates) to cleave the fatty acids from the lipids and convert them to their sodium salts. The tubes were then tightly sealed with a clean Teflon-lined screw cap and vortexed for 10 seconds and placed in boiling water for 5 minutes at 100 oC. After five minutes the tubes were removed from the boiling water and cooled to about 60 oC after which they were again vortexed for 10 seconds. Once no leakage was observed the tubes were heated for a further 25 minutes after which they were cooled to room temperature with cold tap water. The methylation procedure was performed by adding 2.0 ml of the methylation reagent to the content of each tube to convert the sodium form of the fatty acids into fatty acid methyl esters. The tubes were tightly capped and vortexed for about 10 minutes. Following heating of the tubes in boiling water at 80 oC, the tubes were cooled to room temperature with cold tap water. In order to extract the fatty acid methyl esters from the acidic phase and transfer them into organic phase, 1.25 ml of the extraction reagent was added to each tube and mixed end-to-end in a laboratory rotator for 10 minutes. The aqueous (lower) phase was removed and discarded whilst the top phase was kept in the tube. Three millimetres of the base washreagent was added to the tube, vortexed for 10 minutes and allowed to settle. Portion of the top phase in the tube was transferred into vials, capped and analysed by chromatography (GC) with a Hewlett Packard 5890 series II (GMI Inc. USA) equipped with flame ionization detectors auto-sampler, integrator and computer. The oven, detector and injection port temperatures were set at 200, 300 and 275 oC respectively and the Helium carrier gas flow was kept at 100 ml/min. Before starting the first run, the whole system was checked to ensure that pressure in helium, hydrogen, nitrogen and compressed air cylinder was enough and the screen displayed ready. Major fatty acids were identified by comparison of retention times with those of pure bacterial fatty acid methyl esters standard (Microbial Identification System; TSBA library version 3.9; MIDI, USA). rotary shaker (150 rev min-1). In the first stage, 0.2 g (1.0 gl-1) of silver zeolite was added immediately after inoculation. Samples were taking every 15 minutes for two hours. The concentrations of silver ions over each time period were determined by inductively coupled plasma atomic emission spectroscopy (ICP-AES). In the second stage the inoculated media were incubated at 37 oC in a rotary shaker (150 rev min-1) for two hours before exposing them to 0.2 g of silver-loaded zeolite X (1.0 gl-1). Zeolite X without silver ions was used as control. Cells in the late exponential phase were harvested by centrifugation (6900 × g) at 15 oC for 10 minutes and transferred into culture tubes. The extraction was carried out as follows (Sasser 1990): One millilitre of the saponification reagent was added to the cells in the culture tubes (two replicates) to cleave the fatty acids from the lipids and convert them to their sodium salts. The tubes were then tightly sealed with a clean Teflon-lined screw cap and vortexed for 10 seconds and placed in boiling water for 5 minutes at 100 oC. After five minutes the tubes were removed from the boiling water and cooled to about 60 oC after which they were again vortexed for 10 seconds. Once no leakage was observed the tubes were heated for a further 25 minutes after which they were cooled to room temperature with cold tap water. The methylation procedure was performed by adding 2.0 ml of the methylation reagent to the content of each tube to convert the sodium form of the fatty acids into fatty acid methyl esters. The tubes were tightly capped and vortexed for about 10 minutes. Following heating of the tubes in boiling water at 80 oC, the tubes were cooled to room temperature with cold tap water. In order to extract the fatty acid methyl esters from the acidic phase and transfer them into organic phase, 1.25 ml of the extraction reagent was added to each tube and mixed end-to-end in a laboratory rotator for 10 minutes. The aqueous (lower) phase was removed and discarded whilst the top phase was kept in the tube. Three millimetres of the base washreagent was added to the tube, vortexed for 10 minutes and allowed to settle. Portion of the top phase in the tube was transferred into vials, capped and analysed by chromatography (GC) with a Hewlett Packard 5890 series II (GMI Inc. USA) equipped with flame ionization detectors auto-sampler, integrator and computer. The oven, detector and injection port temperatures were set at 200, 300 and 275 oC respectively and the Helium carrier gas flow was kept at 100 ml/min. Before starting the first run, the whole system was checked to ensure that pressure in helium, hydrogen, nitrogen and compressed air cylinder was enough and the screen displayed ready. Major fatty acids were identified by comparison of retention times with those of pure bacterial fatty acid methyl esters standard (Microbial Identification System; TSBA library version 3.9; MIDI, USA). |
III. RESULTS AND DISCUSSION |
| Methyl esters fatty acids detected by gas chromatography (GC) for E. coli K12W-T (Table 1), for S. aureus NCIMB 6571 (Table 2) and P. aeruginosa NCIMB8295 (Table 3) are presented. The results are presented as percentage concentrations with standard errors. MIDI-FAME analysis produced isolate profiles that were characteristic of the assigned genus as reported in literature [15] – [17]. The nomenclature of the fatty acid profiles obtained is as follows: the number of carbon atoms, followed by a colon, the number of double bonds and then by a position of the first double bond from the methyl end (ω ) end of the molecule. The prefix c or t indicate cis or trans configuration of the double bond, cy – cyclopropane fatty acids, Me – the position of the position of the methyl group from the acid end, and -OH indicates the position of the hydroxyl group from the acid end of the molecule. Branched fatty acids were designed as iso or anteiso, if the methyl branch is one or two carbons from the ω end of the acyl chain Sajbidor [29] and Mrozik et al [16]. Peaks that did not display equivalent chain length values of known fatty acids were left unnamed. |
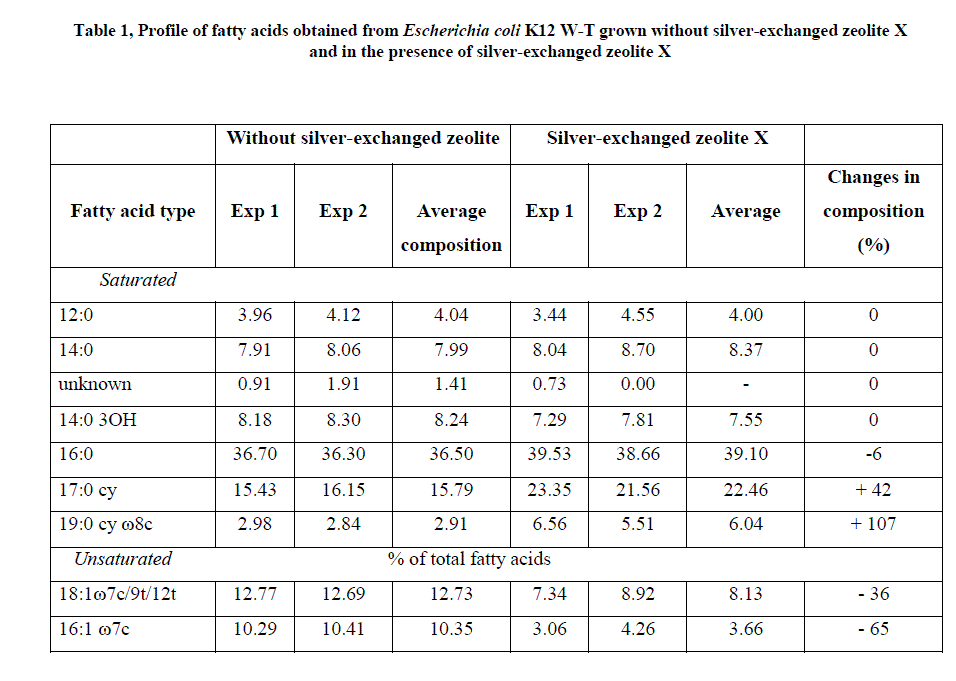 |
| For the interpretation of the obtained results, all isolated fatty acids were grouped into; saturated and unsaturated fatty acids [15, 16]. The first group included four subgroups: straight chain, hydroxyl, cyclopropane and branched fatty acids (iso or anteiso). When cells were challenged with zeolite X (Table 1), the fatty acid composition of E. coli K12 W-T exhibited low proportion of an unknown fatty acid, 19:0 cy ω 8c of the saturated fatty acid obtained, a result similar to that of fatty acids extracted from the cells. These compositions were unaffected upon addition of silver-loaded zeolite X. High proportion of 16:0 fatty acids was obtained when cells were challenged with zeolite X which increased marginally upon addition of silver-loaded zeolite X. The total level of saturated fatty acids in E. coli with zeolite X was 76.9 % whereas it was 88.5 % for silver-loaded zeolite X. The unsaturated fatty acids detected in E. coli K12 W-T were 18:1 7c/9t/12t and 16:1 ω 7c at levels of 12.7 % and 10.4 % respectively. The levels however, dropped to 8.1 % and 3.67 % respectively when challenged with silver-loaded zeolite X. |
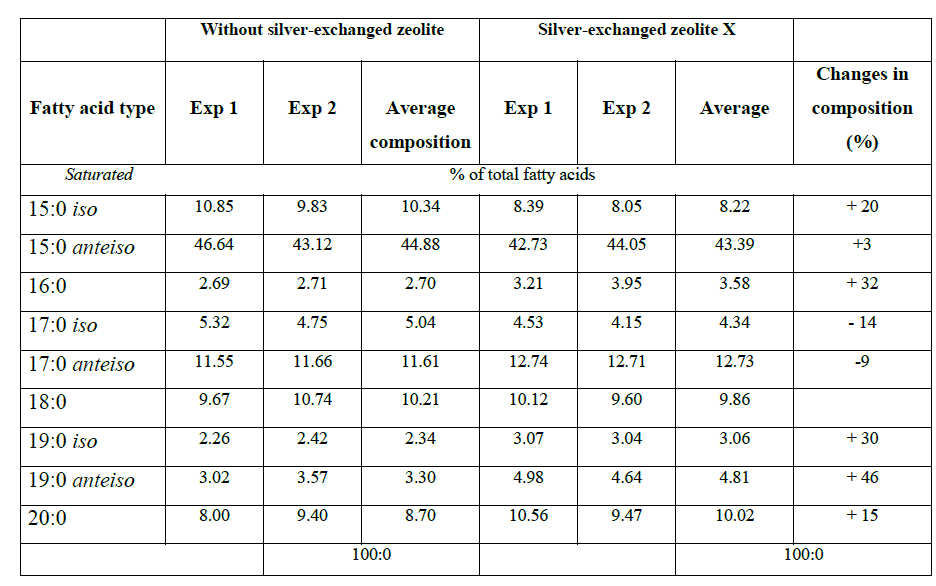 |
| Saturation/unsaturation ratio in E. coli K12 W-T was 3.3 when cells were challenged with zeolite X and 7.4 when cells were challenged with silver-loaded zeolite X. For S. aureus NCIMB 6571 only saturated (branched and unbranched) fatty acids were detected with the characteristics: low proportion of 19:0 iso,16:0, 19:0 anteiso and 17:0 iso; equivalent proportion of 18:0, 15:0 iso and 17:0 anteiso and high proportion of 15:0 anteiso. The results obtained when cells were challenged with silver-loaded zeolite X followed similar trend. P. aeruginosa NCIMB 8295 profile was characterized by high proportion of 16:0 and equivalent proportion of 17:0 cy 16:1 ω 7c fatty acids. In addition, the profile was characterized by low proportion of straight-chain 2-OH and 3-OH fatty acids as well as 18:0, 19:0 cy ω 8c and 12:0 fatty acids. Remarkable changes were observed in the 10:0 3OH, 14:0, and 17:0 cy fatty acids when cells were challenged with silver-loaded zeolite X. A new fatty acid, 18:1 ω 9c was also detected. The saturation/unsaturation ratio was found to be 4.6 for cells challenged with zeolite X and 3.7 for cells challenged with silver-loaded zeolite X. |
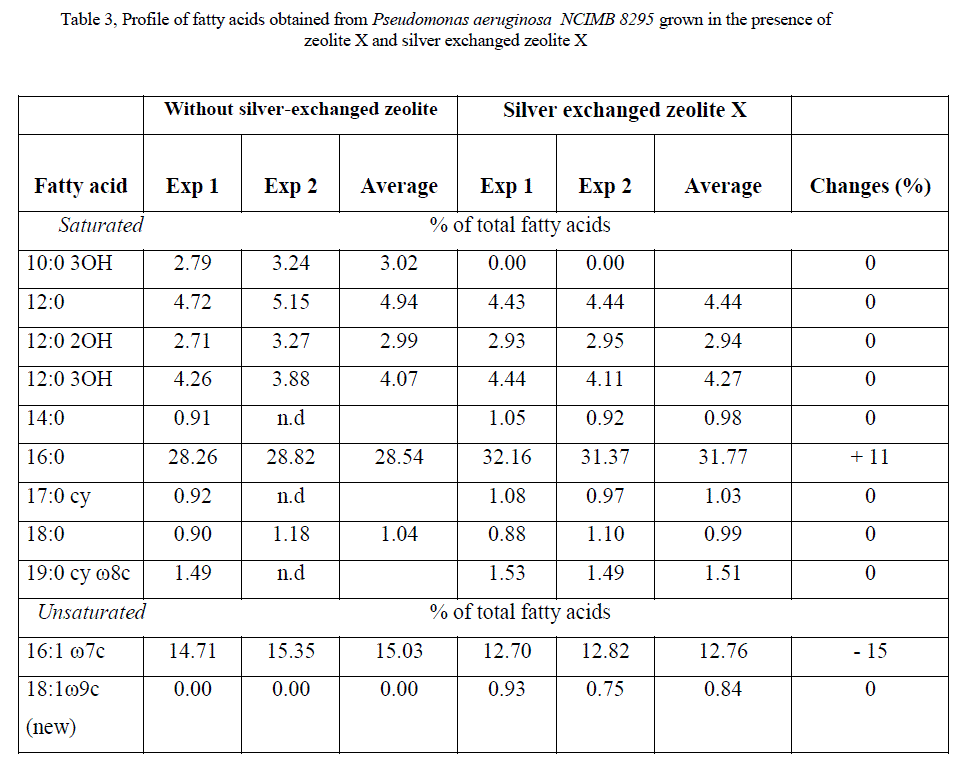 |
| The composition of fatty acids extracted from strains with no zeolite and with zeolite X (zeolite X with no silver ions) were fairly equal but differed with that obtained composition with the profiles obtained with silver-loaded zeolite X. This suggests that changes in fatty acids composition were primarily due to the action of silver ions that were released from the zeolite framework. Silver ions released from the zeolites have been reported to be effective against a wide range of microorganisms and appear to have multiple target sites of microorganisms [3, 26, 32]. The saturated/unsaturated ratio in E. coli K12 W-T increased on exposure to silver-exchanged zeolite X. Similar results were obtained by Mrozik et al [16] when they detected an increase in cyclopropane fatty acid composition in response to naphthalene). Exposure to silver-exchanged zeolite X also changed the composition of branched and hydroxyl fatty acids in E. coli K12 W-T and P. aeruginosa NCIMB 8295. In S. aureus NCIMB 6571 however, all the fatty acids detected were saturated with an increase in the composition of branched fatty acids. Tsitko et al [35] obtained similar results when he studied the impact of different aromatic compounds on whole cell fatty acid composition of Rhodococcus opacus strains GM-14, GM-24 and 1CP. Our results showed a decrease in the composition of 14:0, 10:0 3OH and 17:0 cy fatty acids in P. aeruginosa NCIMB 8295 when their cells were challenged with silver-loaded zeolite X. Gram-positive bacteria cell wall contains three to twenty times more peptidoglycan than their gram-negative counterpart [3, 11]. Since peptidoglycans are negatively charged, it binds some portion of silver. Consequently more silver ions reach the plasma membrane of gram-negative than gram positive bacteria. In addition, gram-negative bacteria have been reported to contain lipids in the cell wall as well as in the cytoplasmic membrane [13]. Fatty acids of cytoplasmic origin are mostly cyclopropane fatty acids. These were detected in both E. coli K12 W-T and P. aeruginosa NCIMB 8295. The absence of cyclopropane fatty acid in S. aureus NCIMB 6571 suggests that all fatty acids were in the peptidoglycan. Hence, factors that influence their susceptibility to antimicrobial agents are likely to be more complex than those of gram-positive bacteria. The saturated/unsaturated ratio in P. aeruginosa NCIMB 8295 decreased from 4.6 to 3.7. Mrozik et al [15] obtained similar results of fatty acid profiles for P. aeruginosa when they studied the influence of naphthalene on fatty acid composition of Pseudomonas sp. JS 150. Cells of E. coli contained significant proportion of 17:0 cy and 19:0 cy ω 8c. Another observation in the fatty acids composition was the appearance of new fatty acid in S. aureus NCIMB 6571 (18:1 ω 9c, Table 2). |
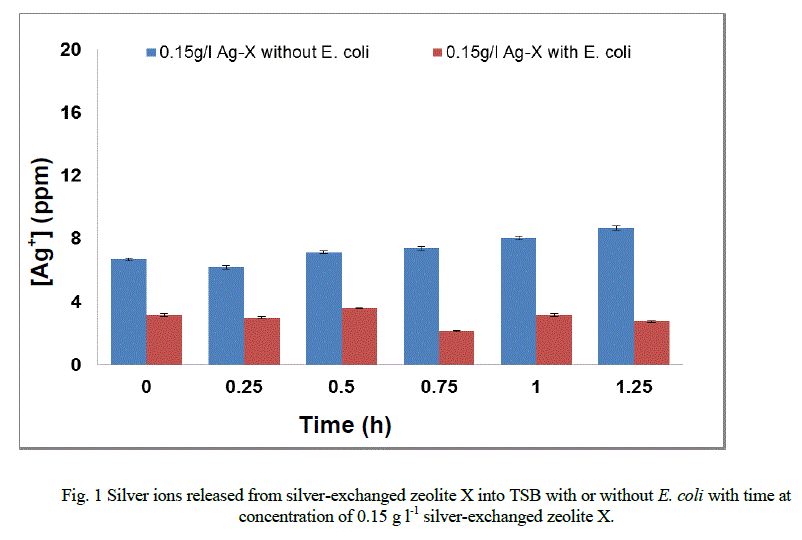 |
| The mechanism of formation of new fatty acids has not been documented (Gutierrez et al., 1999; Mrozik et al. 2004a). However, they may be involved in the protection of strains tested against disruptions of the membrane-cell wall structure [7, 8, 16]. Interaction of silver ions with the fatty acid composition of E. coli K12 W-T S. aureus NCIMB 6571 and P. aeruginosa NCIMB 8295 resulted in significant changes in the composition. These changes are likely to affect the charge, hydration and conformity of their membrane lipids with a consequent changes in the phase behaviour of the lipids [13, 18]. In Figure 1 (a – c), the release profiles of silver again varied from strain to strain on their exposure to 1.0 g l-1 in TSB. In the presence of E. coli (Figure 1a) the release profile of silver ions increased between 0 and 0.15 hours followed by a progressive decrease in the release of silver ions until the end of the exposure period. |
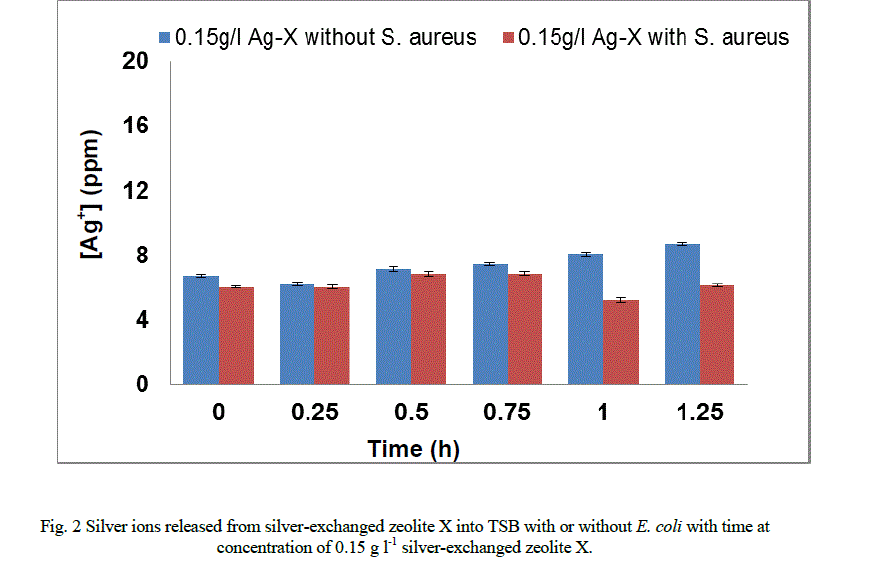 |
| In the presence of S. aureus (Figure 1b), however the release profile of silver ions increased in a similar manner between 0 and 0.15 hours followed by a decrease in release between 0.75 and 1.25 hours. For P. aeruginosa (Figure 1c), the release in silver ions was progressive from 0 to 0.75 hours and decreased progressively until the end of the exposure period. The results of this study have shown that the amount of silver ions needed to effect antimicrobial activity is dependent of the cell structure of the bacteria. As samples were withdrawn from the flask silver ions in the culture were reduced. This implies that more silver needed to be released from the zeolite framework to replace what was removed. According to Le Chatelier’s principle the position of the equilibrium will shift to the right hence more silver ions were likely to be released from the zeolite framework. However, since the release of silver ions is controlled by the zeolite framework, the amount of silver ions released at any time depends on the location of ions in the framework. Silver ions at exchangeable sites are normally more difficult to displace than those at the surface or in the pores[38]. It is likely that this observation might account for the anomalous trend of the silver release from the zeolite framework into the TSB. The uptake of silver ions in bacteria depends on the type of strain being inhibited or killed [5]. Currently, information available on the release of silver ions from silver compounds is very little since results reported in literature are conflicting [2]. While results showed that the amount of silver ions released to effect antimicrobial activity has been found to be directly proportional to the rate of kill in vitro [20, 21, 31, 34], Ovington, [22] Parsons et al [23] showed the contrary. The results obtained from this study showed that silver ions were released from the zeolite in an anomalous trend and inhibition of growth rate. The discrepancies between results have been attributed to the lack of detail on how some tests were performed and the established chemical principles of solubility [2]. As an example it has been shown that a hydrofiber dressing containing silver and carboxymethylcellulose (Ag/CMC) released 0.8 ppm of silver in water and in saline but released 85 ppm of silver in thiosulfate [23, 36]. Nanocrystalline silver released 50 ppm or 70 ppm in water (depending on experimental conditions), 0.8 ppm in saline for the same analysis and 640,000 ppm in thiosulfate [23, 36]. Hence, extent of silver elution depends on the nature of the type of media and the experimental conditions. |
IV CONCLUSION |
| The first objective of this work was to determine whether a relationship exists between the fatty acid composition and susceptibility of bacteria to silver ions released in a controlled manner from a zeolite framework. The saturated/unsaturated of ratio of fatty acids detected in cells grown in the presence of silver-loaded zeolite X decreased for P. aeruginosa NCIMB 8295 but increased for E. coli K12 W-T compared with zeolite X and without any zeolite. This indicates that silver ions released from the zeolite framework exerted substantial stress on the lipid of the bacterial cells. However, there was no strict relationship between the individual fatty acids and susceptibility. The saturated/unsaturated of ratio of fatty acids detected in cells grown in the presence of silver-loaded zeolite X decreased for P. aeruginosa NCIMB 8295 but increased for E. coli K12 W-T compared with zeolite X without silver. |
References
|