e-ISSN: 2319-9849
e-ISSN: 2319-9849
Zhang YL1*, Zhang Y1, Wang X2, Wang S1 and Qiao Y1
1School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing, China
2School of Traditional Chinese Medicine, Capital Medical University, Beijing, China.
Received date: 28/07/2015; Accepted date: 10/08/2015; Published date: 13/08/2015
Visit for more related articles at Research & Reviews: Journal of Chemistry
In contrast to sweet and umami taste, which evolved to recognize a limited subset of nutrients, bitter taste has the onerous task of preventing the ingestion of a large number of structurally distinct toxic compounds. 25 taste receptor type 2 members (T2Rs) have been shown to function as bitter taste receptors. While there is an important question in taste research is how 25 receptors of the human T2Rs family detect thousands of structurally diverse compounds. In silico modeling of T2Rs allowed us to visualize the putative mode of various interactions between agonists and hT2Rs. In this study, ligand-based characterization of the structure function relationship for hT2Rs have been used and the pharmacophore models of T2R1, T2R10, T2R14 and T2R46 have been generated to understand the molecular basis underlying the broad tuning and selectivity of T2Rs members. Moreover, we served T2Rs as representation of Bitter Flavor and verified the relationship between them using virtual screening methods, which also show the scientific variety of Bitter Flavor because of the structural characteristics. The results show that T2Rs agonist pharmacophore models have ability to accumulate bitter herbs and identify the effective components from bitter herbs. It also shows that the Bitter Flavor theory of TCM holding various scientific content because of the structural characteristics. The method used in this paper provides a way for exploring the scientific connotation of five flavors theory of TCM, it can be extended to other TCM theory to solve similar problems.
Five Flavors, Bitter, T2Rs, Pharmacophore, Pharmacological Efficacy.
TCM: Traditional Chinese Medicine; GPCR G: Protein Coupled Receptor; T2Rs: Taste Type?Receptors; TCMD: Traditional Chinese Medicine Database; DS4.0: Accelrys Discovery Studio 4.0; T2R1: Taste receptor type 2 member 1; T2R10: Taste receptor type 2 member 10; T2R14: Taste receptor type 2 members 14; T2R46: Taste receptor type 2 members 46.
Bitter taste is mediated by a family of 25 highly divergent GPCRs (G Protein Coupled Receptors) [1,2]. A large number of T2Rs have been shown to function as bitter taste receptors in heterologous expression assays [3-6], and several have distinctive polymorphisms that are associated with significant variations in sensitivity to selective bitter tastants in mice, chimpanzees and humans [7,8]. An important question in taste research is how 25 receptors of the human T2R family detect thousands of structurally diverse compounds. An answer to this question may arise from the observation that T2Rs in general are broadly tuned to interact with numerous substances. In silico modeling of T2Rs allowed us to visualize the putative mode of various interaction between agonists and hT2Rs (human taste type?receptors).
Despite recent progress in structure determination of GPCRs, structural data on GPCRs are scarce and crystallized receptor proteins exhibit only low amino acid sequence similarity with T2Rs. Therefore, ligand-based characterization of the structurefunction relationship for this GPCR family is necessary to understand the molecular basis underlying the broad tuning and selectivity of its members. Moreover, the pharmacological properties of hT2Rs are characterized by two important features: (i) a rather broad tuning, exemplified by the fact that, based on current information on 20 deorphaned receptors, some hT2Rs responded to up to one-third of all bitter compounds tested [9] and (ii) although highly variable, the average affinity for bitter agonists is rather low compared with other GPCR-ligand interactions [9]. Nevertheless, hT2Rs can discriminate even among chemically very similar bitter compounds with high accuracy [10]. The combination of these two features Results in the manifestation of agonist spectra, which are unique for every single hTAS2R, although some overlaps for individual bitter compounds are evident
According to the Bitter Database built in 2012 (http://bitterdb.agri.huji.ac.il/dbbitter.php), taste type? receptors and specific ligands distribution have been summarized in Table 1, which remind us of some hints about the law of T2Rs. In this study, pharmacophore model of T2R1, T2R10, T2R14 and T2R46 which have been reported possessing more ligands have been generated to study its diversified structural T2Rs.
| BitterDB Receptor ID | Short Name | Organism | Protein Name | Number of Ligands |
|---|---|---|---|---|
| 1 | T2R1 | Human | Taste receptor type 2 member 1 | 35 |
| 2 | T2R3 | Human | Taste receptor type 2 member 3 | 1 |
| 3 | T2R4 | Human | Taste receptor type 2 member 4 | 22 |
| 4 | T2R5 | Human | Taste receptor type 2 member 5 | 1 |
| 5 | T2R7 | Human | Taste receptor type 2 member 7 | 6 |
| 6 | T2R8 | Human | Taste receptor type 2 member 8 | 3 |
| 7 | T2R9 | Human | Taste receptor type 2 member 9 | 3 |
| 8 | T2R10 | Human | Taste receptor type 2 member 10 | 31 |
| 9 | T2R13 | Human | Taste receptor type 2 member 13 | 2 |
| 10 | T2R14 | Human | Taste receptor type 2 member 14 | 47 |
| 11 | T2R16 | Human | Taste receptor type 2 member 16 | 10 |
| 12 | T2R38 | Human | Taste receptor type 2 member 38 | 21 |
| 13 | T2R39 | Human | Taste receptor type 2 member 39 | 20 |
| 14 | T2R40 | Human | Taste receptor type 2 member 40 | 11 |
| 15 | T2R41 | Human | Taste receptor type 2 member 41 | 1 |
| 16 | T2R42 | Human | Taste receptor type 2 member 42 | 0 |
| 17 | T2R43 | Human | Taste receptor type 2 member 43 | 16 |
| 18 | T2R44 | Human | Taste receptor type 2 member 44 | 8 |
| 19 | T2R45 | Human | Taste receptor type 2 member 45 | 0 |
| 20 | T2R46 | Human | Taste receptor type 2 member 46 | 27 |
| 21 | T2R47 | Human | Taste receptor type 2 member 47 | 10 |
| 22 | T2R48 | Human | Taste receptor type 2 member 48 | 0 |
| 23 | T2R49 | Human | Taste receptor type 2 member 49 | 2 |
| 24 | T2R50 | Human | Taste receptor type 2 member 50 | 2 |
| 25 | T2R60 | Human | Taste receptor type 2 member 60 | 0 |
Table 1: Taste TypeⅡ Receptors and Specific Ligands Distribution[11].
As a guide principle, TCM property theory has played an important role in syndrome differentiation and clinical prescription for thousands of years [11-13]. The theory of Five Flavors which has been used to summarize the function of drugs is one of the native-born medicine theories in China. According to TCM property theory, the favour of bitter is one of the five basic tastes (the other four are sweetness, sourness, saltiness, and bitterness). In this study, we served Taste Type?Receptors (T2Rs) as one of representations of Bitter Flavor and try to verify the relationship between them. In this study, we have also collected some natural agonists of T2Rs as well as the TCM sources and the related properties (Table 2).
| Compounds | TCM Source | Herbal Source | Property | T2Rs |
|---|---|---|---|---|
| Benzoin | An Xi Xiang | Styrax tonkinensis (Pierre) Craib ex Hart. | Bitter/Pungent, Normoal | T2R10, T2R14 |
| Andrographolide | Chuan Xin Lian | Andrographis paniculata (Burm. f.) Nees | Bitter, Cold | T2R46, T2R47, T2R50 |
| Sinigrin | Ting Li Zi | Lepidium apetalum Willd. | Pungent/Bitter, Cold | T2R16, T2R38 |
| Xanthohumol | Pi Jiu Hua | Humulus lupulus L. | Bitter/Pungent, Cool | T2R1, T2R14, T2R40 |
| Adhumulone | Pi Jiu Hua | Humulus lupulus L. | Bitter/Pungent, Cool | T2R1, T2R40 |
| Adlupulone | Pi Jiu Hua | Humulus lupulus L. | Bitte/Pungent, Cool | T2R1, T2R14 |
| Absinthin | Ku Ai | Artemisia absinthum L. | Bitter/Pungent, Warm | T2R10, T2R14, T2R46, T2R47 |
| Quassin | Ku Shu Pi | Celastrus angulatus Maxim. | Cold, Bitter | T2R4, T2R10, T2R14, T2R46, T2R47 |
| Amarogentin | Ku Xing Ren | Prunus armeniaca L. var. ansu Maxim. | Bitter, Warm | T2R1, T2R4, T2R16, T2R39, T2R43, T2R46, T2R47, T2R50 |
| Quinine | Jing Ji Na | Cinchona ledgeriana Moens | Bitter, Cold | T2R4, T2R7, T2R10, T2R14, T2R39, T2R40, T2R43, T2R44, T2R46 |
| Aloin | Lu Hui | Aloe barbadensis Miller | Bitter, Cold | T2R43, T2R44 |
| Aristolochic acid | Ma Dou Ling | Aristolochia contorta Bge. | Bitter/Pungent, Cold | T2R14, T2R43, T2R44 |
| Brucine | Ma Qian Zi | Strychnos nuxvomica L. | Bitter, Cold | T2R4, T2R46 |
| Picrotoxinin | Mu Fang Ji | Coculus trilobus (Thunb.) DC. | Bitter/Pungent, Cold | T2R10, T2R14, T2R46, T2R47 |
| Strychnine | Ma Qian Zi | Strychnos nuxvomica L. | Bitter, Cold | T2R10, T2R46 |
Table 2: Natural agonists of T2Rs and TCM sources [11].
A pharmacophore model can be considered as the highest common denominator of a group of molecules exhibiting a similar pharmacological profile and which are recognized by the same site of the target protein. HipHop algorithm, which attempts to produce an alignment of compounds expressing certain activity against a particular target and by superposition of diverse conformations to find common three-dimensional arrangements of features shared between them [2], has been used to build pharmacophore of T2R1, T2R10, T2R14, and T2R46. The best pharmacophore model of T2Rs have been built to search Traditional Chinese Medicine Database (TCMD) (Version 2009), questing for the agonists from traditional Chinese medicine for further research of the connection between the “Bitter Flavor” and T2Rs.
Compounds and Biological DataThe studies were implemented on a series of T2R1 agonists reported by literature [11,14-16]. The structures of agonists are listed in Figure 1. Considering the distribution of structural diversity, twelve compounds were selected to generate the pharmacophore model and the other compounds were used as test set to validate the model. The agonists of T2R10, T2R14 and T2R46 have been listed in Supplementary Materials.
Pharmacophore generation and validation:In this study, the main steps of pharmacophore generation are as follows [17-18]:
Conformational analysisThe 3D qualitative pharmacophore hypotheses have been constructed by HipHop (Common Feature Pharmacophore Generation) within Accelrys Discovery Studio 4.0 (DS4.0). Ligand conformations were created within the relative energy threshold of 20 kcal/mol by the BEST mode (Best Quality Conformer Generation) at the number of 255 maximum conformations.
Common feature mappingAccording to the Feature Mapping’s initial analysis, hydrogen bond acceptor (A), hydrogen bond donor (D), hydrophobic (H), and ring aromatic (R) which well-mapped all of the training set ligands, has been selected during the pharmacophore generation. These pharmacophore features can be characterized the interaction between the ligand and receptor.
Pharmacophore generation and verificationThe HipHop algorithm attempts to produce an alignment of compounds by superposition of diverse conformations to find common three-dimensional arrangements of features shared between them. The Minimum Features has been set as 3, while the Maximum Features was 10. And we set the maximum number of pharmacophores was 10.
In this study, for validating the pharmacophore hypotheses using external test set molecules, which have not been used for pharmacophore model generation, a test database of Experimentally known T2Rs agonists embedded in a database consisting of some drug-like molecules (taken from the MDL MDDR database: Version2007.2) was constructed to evaluate all of the pharmacophore models. All of the identified ligands were filtered by Lipinskis Rule and were similar in chemical structural characteristics. To evaluate the performance of the models, four parameters (i.e., A%, Y%, N, and CAI) and the relationship between them was revealed in Figure 2 with the correlativity as follows [17]:
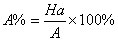 (1)
(1)
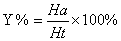 (2)
(2)
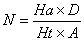 (3)
(3)
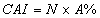 (4)
(4)
A% can represent the ability to identify active compounds from the test database and Y% represents the proportion of active compounds in hit compounds. N, the index of effective identification, is used to evaluate the ability of the models to identify active compounds from non-active compounds. CAI, a comprehensive evaluation index, is used to identify the best pharmacophore model. D is the total number of compounds in the test database and A is the number of active compounds. Ht is the total number of hit compounds from the test database and Ha is the number of active hit compounds from the test database. The model with the highest value of CAI is considered to be the best.
Virtual ScreeningAccording to the performance in terms of the enrichment factor (CAI value) of pharmacophore models, the most excellent pharmacophore model can be useful filters for virtual screening to identify T2R1, T2R10, T2R14 and T2R46 agonists within large compound repositories in TCM. This research select the model with highest CAI value served as a query to perform 3D Flexible Searching operation in DS 4.0 to search Traditional Chinese Medicine Database (TCMD, version 2009), which contains 23033 natural compounds from 6735 medicinal plants. Moreover, all potential hit compounds in the database should be satisfied the Lipinski’s rule of five requirements [19-20].
Twelve compounds were used as the training set for a HipHop running. The top 10 pharmacophore models with the calculation Results are detailed in Table 3. According to the simulated Results, the main features contains, Hydrophobic (H) and H-Bond Acceptors (A) have been generated. Both of the Direct Hi and Partial Hit values of these 10 pharmacophore models are “111111” and “000000” which confirm that all of the eleven molecules in training set have been taken into the generation of the models. The “4” value of Max Fit proves to us that all of the features in models can match with the molecules in training set. The Rank scores indicate the matching degree between the pharmacophore feature and the molecules. In general, the higher the score, the model matched more satisfactorily. The pharmacophore model calculation Results of T2R10, T2R14 and T2R46 have been listed in Supplementary Materials.
| Model | Features | Rank | Direct Hit | Partial Hit | Max Fit |
|---|---|---|---|---|---|
| 01 | HHHA | 50.372 | 111111 | 000000 | 4 |
| 02 | HHHA | 50.260 | 111111 | 000000 | 4 |
| 03 | HHHA | 49.553 | 111111 | 000000 | 4 |
| 04 | HHHA | 49.285 | 111111 | 000000 | 4 |
| 05 | HHHA | 49.096 | 111111 | 000000 | 4 |
| 06 | HHHA | 48.796 | 111111 | 000000 | 4 |
| 07 | HHHA | 48.796 | 111111 | 000000 | 4 |
| 08 | HHHA | 48.261 | 111111 | 000000 | 4 |
| 09 | HHHA | 48.105 | 111111 | 000000 | 4 |
| 10 | HHHA | 47.887 | 111111 | 000000 | 4 |
Table 3: T2R1 Pharmacophore model calculation results.
Pharmacophore Models ValidationOn the basis of the diagram of indicators evaluating the pharmacophore models showed before, we have shown the parameter values for each pharmacophore model in Table 4. NO.1 MODEL with the highest CAI value has been selected to screen TCMD2009 database. The pharmacophore feature of Model 1 mapped with ligand Xanthohumol has been showed in Figure 3. The parameter values for T2R10, T2R14 and T2R46 pharmacophore models have been listed in Supplementary Materials.
| Model | Aa | Db | Htc | Had | A%e | Nf | CAIg |
|---|---|---|---|---|---|---|---|
| 1 | 27 | 180 | 41 | 19 | 70.4 | 3.09 | 2.17 |
| 2 | 27 | 180 | 46 | 17 | 63.0 | 2.46 | 1.55 |
| 3 | 27 | 180 | 46 | 18 | 66.7 | 2.61 | 1.74 |
| 4 | 27 | 180 | 48 | 17 | 63.0 | 2.36 | 1.49 |
| 5 | 27 | 180 | 42 | 16 | 59.3 | 2.54 | 1.50 |
| 6 | 27 | 180 | 45 | 18 | 66.7 | 2.67 | 1.78 |
| 7 | 27 | 180 | 47 | 18 | 66.7 | 2.55 | 1.70 |
| 8 | 27 | 180 | 46 | 18 | 66.7 | 2.61 | 1.74 |
| 9 | 27 | 180 | 49 | 17 | 63.0 | 2.31 | 1.46 |
| 10 | 27 | 180 | 59 | 16 | 59.3 | 1.81 | 1.07 |
Aa is the number of active compounds; Db is the number of compounds in the test database; Htc is the number of hits using pharmacophores to search; Had is the number of active hits using pharmacophores to search; A%e represents the ability to identify active compounds from the test database; Nf represents the ability to identify active compounds from nonactive compounds; CAIg is the comprehensive appraisal index.
Table 4: T2R1 Parameter values for each pharmacophore model.
To quest for the potential agonists of T2R1, T2R10, T2R14 and T2R46, the pharmacophore model generated by HipHop was used as the query to perform a search of all of the known compounds from TCMD2009. According to the Results of virtual screening, for example in T2R1, 186 compounds showing notable pharmacological activities have been hit, were documented in 57 Chinese Herbs from Chinese Pharmacopoeia 2010. 53.7% hits belong to the flavor of bitter (Table 5). No doubt target T2R1 bear some relation to “bitter flavor”.
| Ligands | TCM Souces | Flavors | T2Rs |
|---|---|---|---|
| TCMD059 | Bai Guo | Sweet/Bitter | T2R10,T2R14 |
| TCMD060 | Bai Guo | Sweet/Bitter/ | T2R10,T2R14 |
| TCMD061 | Bai Shao | Bitter/Sour | T2R10,T2R14 |
| TCMD062 | Bai Shao | Bitter/Sour | T2R10,T2R14 |
| TCMD063 | Chai Hu | Bitter | T2R10,T2R14,T2R46 |
| TCMD064 | Chai Hu | Bitter | T2R10,T2R14,T2R46 |
| TCMD065 | Chai Hu | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD066 | Chai Hu | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD067 | Chuan Xin Lian | Bitter | T2R10,T2R14,T2R46 |
| TCMD068 | Chuan Xin Lian | Bitter | T2R10,T2R14,T2R46 |
| TCMD069 | Da Huang | Bitter | T2R10,T2R46 |
| TCMD070 | Da Huang | Bitter | T2R10,T2R14 |
| TCMD071 | Da Huang | Bitter | T2R10,T2R14,T2R46 |
| TCMD072 | Da Huang | Bitter | T2R1,T2R10,T2R46 |
| TCMD073 | Da Huang | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD074 | Da Huang | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD075 | Dan Sheng | Bitter | T2R10,T2R46 |
| TCMD076 | Dan Sheng | Bitter | T2R10,T2R46 |
| TCMD077 | Dan Sheng | Bitter | T2R10,T2R14,T2R46 |
| TCMD078 | Dan Sheng | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD079 | Dan Sheng | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD080 | Huang Lian | Bitter | T2R46 |
| TCMD081 | Huang Qin | Bitter | T2R10 |
| TCMD082 | Huang Qin | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD083 | Lian Qiao | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD084 | Lian Qiao | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD085 | Long Dan | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD086 | Ma Qian Zi | Bitter | T2R1,T2R10,T2R46 |
| TCMD087 | Zhe Bei Mu | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD088 | Zhe Bei Mu | Bitter | T2R1,T2R10,T2R14,T2R46 |
| TCMD089 | Zhi Qiao | Bitter/Pungent/ Sour | T2R1,T2R10,T2R46 |
Table 5: Partial Virtual Screening Hits of T2Rs Agonists.
According to the simulation Results, the “hit” compounds are derived from medicinal herbs sharing bitter flavor, such as Salvia miltiorrhiza Bunge, Coptis chinensis Franch and Rheum palmatum L. Moreover, the number of “Hit” compounds whose Hit Score higher than 80.0 is 68 and 51.5% compounds are from pungent TCM sources. It indicated that the pharmacophore model of T2Rs can gather the same structural characteristics of pungent TCM. The model is capable to identify the compounds drawing from bitter TCM.
Method Limitation Analysisthis paper, 4 bitter taste receptor were studied through pharmacophore model and virtual screening to explore its relationship with bitter flavor. However, in addition to these 4 targets, the T2Rs family includes 21 other GPCRs. Therefore, this study is not enough to reveal the relationship between T2Rs family and bitter property, and further study is also needed to interpret bitter flavor through T2Rs family.
In this paper, ligand-based characterization of the structure-function relationship for hT2Rs has been used and the pharmacophore model of T2R1, T2R10, T2R14 and T2R46 have been generated. The Results show that T2Rs agonist pharmacophore models have ability to accumulate bitter herbs and identify the effective components from bitter herbs. It also show that the Bitter Flavor of TCM holding various scientific content because of the structural characteristics The method used in this paper provides a way for exploring the scientific connotation of five flavors theory of TCM, it can be extended to other TCM theory to solve similar problems.
Y-J Qiao and Y-L Zhang have been conceived and designed the experiments. Y-X Zhang and X Wang have been involved in processing data and preparing the manuscript. S-F Wang participated in the Discussion of views in the paper. All authors have read and approved the final manuscript.
This study was financially supported by the National Science Foundation of China (Project No. 81430094)